Abstract
Despite Paraquat (PQ) being banned in several countries, it is still one of the most commonly used herbicides in agriculture. This compound is known to induce damaging effects on human and animal brain cells by generating Reactive Oxygen Species (ROS). However, there is few evidence of PQ effect on Human Brain Microvascular Endothelial Cells (HBMECs), one of the major component of the Blood–Brain Barrier (BBB). The present study aimed at unraveling biological mechanisms associated to the exposure of 1, 10 and 100 µM of PQ for 24 h on HBMECs. High-throughput mass spectrometry-based proteomics using data-independent acquisition (DIA) was applied. Biological pathway enrichment and cellular assays such as mitochondrial respiration and cholesterol level were performed to verify proteomics results. A total of 3753 proteins were quantified out of which 419 were significantly modulated by paraquat exposure. Biological pathway enrichment revealed the ubiquinone metabolism, a pathway directly linked to mitochondrial complex I proteins, confirming the well-known mechanism of PQ inducing oxidative stress. Additionally, this study also described the cholesterol biosynthesis modulation on HBMECs not yet described. In conclusion, our data indicate the toxic effect of PQ on HBMECs by downregulating proteins involved in mitochondrial complex I and cholesterol pathways.
Similar content being viewed by others
Introduction
Epidemiological and toxicological studies have reported an association between environmental toxicant exposure and neurodegenerative diseases1,2,3,4,5. Among these environmental toxicants, paraquat (PQ) is one of the most widely used herbicide in many parts of the world. Nevertheless, PQ is also known for its acute toxicity and chronic effects3,6. Agricultural workers and farmers, who are not sufficiently protected, are still applying this chemical and increasing risk of developing adverse health effects such as respiratory system default, reproductive problems or increasing about twofold their risk of developing Parkinson’s disease4,5,7,8,9,10,11,12,13. Because of these evidences, PQ was banned by the European Court of First Instance in several countries, mostly in the European Union countries14. However, despite the known public health issues of PQ, it is still applied by millions of agricultural workers in about 100 countries, mainly the global south countries but also by the United States. Moreover, data from US Geological Survey’s Pesticide National Synthesis Project pointed out that use of PQ doubled over the past decade.
The toxic mode of action of PQ is to induce oxidative stress through the generation of Reactive Oxygen Species (ROS), creating a cell redox imbalance1,15,16. In this redox relation, interaction of PQ with mitochondria remains an important and one of the most described aspect of its toxicity, particularly in brain17,18. Mitochondrial complex I and III activities are the two main complexes from the electron chain transport impacted by PQ exposure. Their alteration lead to the inhibition and dysfunction of mitochondrial function17,18,19. Despite the presence of the blood–brain barrier (BBB), a unique anatomical and physiological barrier between the bloodstream and the extracellular space of the brain, it was demonstrated that PQ can be transported into the brain by neutral amino acid transport system6,20. This toxin is also eliminated much more slowly in the brain than any another organ such as the liver, suggesting longer-lasting effects in the brain.
HBMECs are different from other endothelial cells due to their unique phenotype21,22. They possess specialized junctional complexes such as tight, adherens or gap junctions. These junctions are able to maintain the BBB integrity23,24 by connecting HBMECs together. Thereby, they prevent compounds from paracellular diffusion across the BBB25,26,27. The existence of specific metabolic transporters (e.g. solute carrier and ATP-binding cassette family) also control the movement of nutrients, ions, toxins or xenobiotics across the endothelia21. Moreover, HBMECs are determined by intimate contact with other members of the neurovascular unit (NVU) such as neurons, astrocytes, microglia, pericytes and extracellular matrix molecules27. These complex interactions contribute to the dynamic regulation of microvascular permeability and regulate the function of HBMECs during normal and abnormal BBB activity28,29,30, explaining their importance in brain research.
Since environmental toxicant are important risk factors of neurodegeneration, it is crucial to put forward mechanisms by which they cause neurotoxicity. The aim of this study was to identify detrimental mechanisms caused by PQ on HBMECs. Data-independent acquisition mass spectrometry (DIA-MS) was applied to explore proteome profiles and biological pathways impacted by PQ at 1, 10 and 100 µM for 24 h and to demonstrate its role as an oxidative stress inducer on HMBECs. Proteomics and biological pathway enrichment analysis provide evidence of the modulation of a well-known pathway—the ubiquinone metabolism—linked to the mitochondrial complex I proteins. More is being learned about the effects of exposure to PQ on HBMECs as results provide novel insight on the alteration of cholesterol biosynthesis.
Results
MTS proliferation and LDH cytotoxicity assays of human brain microvascular endothelial cells exposed to paraquat
PQ-induced toxicity was evaluated in HBMECs for 24 h. MTS proliferation assay indicated that PQ concentrations above 100 µM were significantly decreasing cell proliferation (Supplementary Fig. S1). Regarding cytotoxicity, PQ concentration above 100 µM was significantly increasing cellular death indicating its toxic effect (Supplementary Fig. S1). These results suggested that PQ concentration at 0.1, 1, 10 and 100 µM did not affect cell proliferation and had no cytotoxic effect on HBMECs. Moreover, other in vitro and in vivo studies applying PQ demonstrated that concentration up to 100 µM can be used31,32,33.
Proteomics and pathway enrichment analysis of human brain microvascular endothelial cells after paraquat exposure
To further elucidate PQ-induced mechanisms, Data Independent Acquisition (DIA)-based proteomics analysis was applied. More than 3500 proteins were quantified for PQ-exposed HMBECs at 1, 10 and 100 µM, respectively (Fig. 1a).
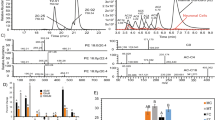
(a–f) Proteomics data and enrichment pathway results of PQ-treated HBMECs at 1 µM, 10 µM and 100 µM for 24 h. (a) Venn diagram displaying a comparison of total quantified proteins in HBMECs after PQ concentrations at 1, 10 and 100 µM. (b) Venn diagram displaying a comparison of total DEPs in HBMECs after PQ concentrations at 1, 10 and 100 µM. (c–e) Volcano plots displaying the distribution of all proteins after PQ concentrations at 1 µM (c), 10 µM (d) and 100 µM (e) in HBMECs. Downregulated proteins are in blue. Upregulated proteins are in red. X axis corresponds to log2(FC), Y axis corresponds to − log10(p-value). (f) Top ten of pathway maps enriched by MetaCore software from the lists of DEPs (FC > 1.2, p-value ≤ 0.05, N = 3) after PQ concentrations at 1 µM (purple), 10 µM (orange) and 100 µM (green) for 24 h. The top 10 pathways for each condition are represented in the graph. X axis corresponds to − log10(p-value), Y axis corresponds to the name of biological pathways and the dashed line represents the enrichment p-value cut-off of 0.05.
Only six proteins were differentially expressed in HBMECs exposed to PQ at 1 µM versus untreated control (Fig. 1b and Supplementary Table S1). Sixty-six proteins were modified by PQ concentration at 10 µM (Fig. 1b and Supplementary Table S2). Finally, 419 proteins were differentially expressed for PQ at 100 µM (Fig. 1b and Supplementary Table S3). These results indicated that the number of differentially expressed proteins (DEPs) is associated to a dose–response manner (Fig. 1c–e). To analyze PQ-effect on HMBECs at the biological pathway level, an enrichment analysis was performed in Metacore software. DEP lists from proteomics experiments were used (Supplementary Tables S1, S2 and S3). Due to the very low number of DEPs in HBMECs exposed to PQ at 1 µM versus untreated control, enrichment pathway was not considered. Pathways enrichment of HBMECs exposed to PQ at 10 µM show highly significant enrichment p-value for the two biological pathways—“SCAP/SREBP Transcriptional Control of Cholesterol and FA Biosynthesis” and ”Cholesterol Biosynthesis ” (p value of 1.61 × 10−08 and 1.11 × 10−07, respectively) (Fig. 1f). Both of these impacted biological pathways were also observed in the top ten of the pathways modified in HMBECs exposed to PQ at 100 µM—“SCAP/SREBP Transcriptional Control of Cholesterol and FA Biosynthesis” (p value = 6.83 × 10−11) and ”Cholesterol Biosynthesis ” (p value = 4.93 × 10−05) (Fig. 1f). However, the second most affected pathway after PQ exposure at 100 µM revealed to be the “Ubiquinone Metabolism” (p value = 2.90 × 10−07) (Fig. 1f).
Ubiquinone metabolism modulation after paraquat exposure on human brain microvascular endothelial cells
Proteomics results as well as pathway enrichment analysis of PQ-exposed HBMECs at 100 µM highlighted a highly altered biological process linked to oxidative stress; the ubiquinone metabolism (Fig. 1). This modulated pathway is implicated in mitochondrial dysfunction, a toxic effect of PQ widely studied mostly in the rat brain15,18,34. Based on pathway enrichment results, the heat map illustrated proteins involved in the ubiquinone metabolism (Fig. 2a).
(a–c) Mitochondrial dysfunction of PQ-treated HBMECs. (a) Heat map of NADH-ubiquinone proteins obtained by MS and altered by PQ exposure. Stars indicate that protein level was modified in HBMECs exposed to PQ for 24 h (LFDR ≤ 0.05; |FC|> 1.2). (b) Time-course experiment to assess mitochondrial stress in HBMECs using different PQ concentrations (1, 10 and 100 μM) and control (water). (c) Basal respiration, spare respiratory capacity, proton leak and ATP production are represented for each concentration. Data are represented as means ± SD of three biological replicates. ** corresponds to p-value ≤ 0.01 and **** corresponds to p-value ≤ 0.0001.
To verify this observation, as NADH-ubiquinone oxidoreductase proteins are involved in the first step of mitochondrial respiration35, a mitochondrial dynamics and bioenergetics assay of PQ-exposed HBMECs was performed. Mitochondrial function assay showed a significant decrease of maximal respiration and spare respiratory capacity for PQ-treated HBMECS at 10 µM and 100 µM (Fig. 2b,c). These results combined to quantitative proteomic results and biological pathway enrichment analysis confirmed that HBMECs exposed to the highest PQ concentration are more susceptible to trigger an oxidative stress-related mechanism.
Cholesterol biosynthesis modulation induced by paraquat on human brain microvascular endothelial cells
Proteomics results as well as pathway enrichment analysis (Fig. 1) suggested that cholesterol biosynthesis is highly altered in HBMECs exposed to PQ at 10 and 100 µM. Fifty-seven DEPs were shared between PQ concentrations at 10 and 100 µM (Fig. 1b). Of those 57 DEPs, a cluster was retrieved as the cholesterol and lipid biosynthetic process (FASN, ACSL3, FADS2, HMGSCS1, DHCR7, IDI1, DHCR24, CYP51A1, MVD, ACAT2, FAR1) (Fig. 3a). The heat map demonstrated that key proteins involved in cholesterol biosynthesis were significantly decreasing as PQ concentration is increasing (Fig. 3a). These crucial proteins were involved in every step of the cholesterol biosynthesis as demonstrated in Fig. 3b, meaning that there is a global impact on cholesterol biosynthesis in HBMECs due to PQ exposure (starting at 10 µM of PQ).
Cholesterol biosynthesis modulation in HBMECs after PQ exposure. (a) Heat map of cholesterol proteins obtained by MS and altered by PQ exposure. Stars indicate that protein level was modified in HBMECs exposed to PQ for 24 h (LFDR ≤ 0.05; |FC|> 1.2). Protein fold-change (FC) were displayed as colours ranging from orange to white as shown in the key. (b) Representation of the cholesterol biosynthesis key steps. Arrows indicate that protein was decreasing in HBMECs exposed to PQ for 24 h (LFDR ≤ 0.05; |FC| > 1.2). Cholesterol measurement of HBMECs exposed to PQ for 24 h. (c) Time-course run of cholesterol measurement using different PQ concentrations (1, 10 and 100 μM), control (water) and positive control (cholesterol 60 μM). Data are represented as means ± SD of three biological replicates. * corresponds to p-value ≤ 0.05, ** corresponds to p-value ≤ 0.01, *** corresponds to p-value ≤ 0,001 and **** corresponds to p-value ≤ 0.0001.
To verify cholesterol biosynthesis modulation after PQ exposure, a cellular assay measuring cholesterol level was performed with three different PQ concentrations (1, 10 and 100 µM) for 24 h, 48 h and 72 h. The cholesterol assay revealed a significant decrease in cholesterol level at 10 µM of PQ for 72 h as well as at 100 µM for 24 h, 48 h and 72 h (Fig. 3c). These results combined to proteomics and enrichment pathway analysis (Fig. 1) supported the hypothesis that HBMECs exposed to PQ could affect the cellular cholesterol biosynthesis.
Discussion
Environmental xenobiotics are still a public concern due to their adverse effects. Indeed, exposure to paraquat (PQ), a non-selective herbicide, has shown some negative effects on agricultural workers such as reduced lung function, skin burn or irritation and eye damage9,36,37. In addition, epidemiological and toxicological studies have also demonstrated a link between paraquat exposure and an increasing risk to develop neurodegenerative diseases1,2,3,38,39. PQ acts by generating superoxide radicals resulting in lipid peroxidation of cellular membranes40. In studies conducted on animal and human brain, it has largely been documented that PQ is able to inhibit the electron transport chain within mitochondria leading to the production of reactive oxygen species (ROS)15,16,18,34,41,42. However, even if PQ toxicity was verified in plethora of research, its use is strongly widespread and often under unsafe and no restricted conditions.
Moreover, many studies have shown that ROS generation is damaging brain cells such as neurons or astrocytes but consequences of PQ exposure in human brain microvascular endothelial cells are still not investigated. This study aimed to explore and give an insight to biological pathways impacted by PQ exposure by using a proteomics quantitative MS-based strategy and enrichment pathway analysis.
PQ concentrations of 1, 10 and 100 µM were selected according to our results from cytotoxicity and proliferation assays and based on several in vitro and in vivo studies using PQ31,32,33,43,44. Indeed, a study using astrocytes and neurons has reported that PQ elicit significant effect at 100 µM after 24 h exposure, supporting PQ concentration chosen in our study as well as the short exposure time of 24 h31. In another research using mouse brain, they selected PQ concentration of 100 µM as the highest one32. However, it is worth noticing that PQ-exposed astrocytes concentration used in the study of Zheng et al. was between 2 to 8-fold higher than in our research45. With this last study, we notice that PQ concentration is cell-type dependent and we highlight the need to be confirmed by cellular assays. Additionally, results from a recent paper from Yuan et al. described that blood concentration of patients after paraquat acute poisoning ranged from 0.10 to 20.62 μg/mL, which correspond to 0.38–80 µM. Nonetheless, our study was performed on a primary cell monolayer culture meaning that interaction effect with other cell types of the BBB (i.e.: glial cells, neurons) is not considered. This interaction may strongly modify and influence brain endothelial cells phenotype as well as PQ concentration to use.
As described in many studies, PQ is causing mitochondrial dysfunction due to the cytotoxic molecule generation such as nitric oxide radicals or other ROS18,34,42,46,47,48,49,50. Similarly, our proteomics results has shown 11 altered proteins involved in the mitochondrial respiratory complex I (NADH-ubiquinone oxidoreductase). Pathway enrichment analysis as well as respirometric assay of mitochondria biogenesis verified these results by indicating an altered pathway linked to complex I of mitochondrial respiration and a statistically significant decrease of mitochondrial function in a dose-dependent manner. These results are consistent with previous studies supporting a deleterious effect of PQ on mitochondria in other cell types18,34,42,46,47,48,49. They are also in line with three studies displaying an association with dysfunction or inhibition of mitochondrial complex I activities5,47,51. Thereby, reduction or alteration in ubiquinone synthesis could impair TCA cycle and mitochondrial respiration, which might change redox environment causing mitochondrial dysfunction. Our findings led to support that PQ is inducing mitochondrial dysfunction also on human brain microvascular endothelial cells.
Furthermore, our proteomics data analysis highlighted the modulation of the cholesterol biosynthesis pathway in PQ-treated HBMECs. Cholesterol biosynthesis, originated from the mevalonate pathway, is vital to ensure fundamental cellular processes52,53,54. Indeed, this sterol is required to reinforce and organize cell membrane as well as to maintain and form lipid rafts52,53,54. Additionally, cholesterol has an essential role by acting as a precursor of steroid hormones, bile acids and oxysterols, and as a regulator of cell signalling52,53,54. In the brain, cholesterol is synthetized in situ, as the plasma lipoproteins are not crossing an intact BBB53,55,56. The first step of mevalonate pathway resulting to the cholesterol synthesis is executed by the transferase, 3-hydroxy-3-methylglutaryl-CoA synthase 1 (HMGCS)57,58. This critical cholesterol biosynthesis enzyme is responsible of the acetyl coenzyme A condensation with acetoacetyl coenzyme A to produce HMG-CoA by a catalytic process57,58. In the present study, this protein was found to be the most significantly decreased, pointing up the first failure in the cholesterol biosynthesis cascade. Smith et al. indicated that reduced expression of HMGCS in mice cerebral cortex is linked to a cholesterol downregulation, supporting our observation59. The mevalonate kinase (MVK), an enzyme involved in the conversion of the mevalonate into the mevalonate-5-phosphate, is the converting enzyme following that of HMGCR. It was also shown to be significantly downregulated in this cascade. Furthermore, one study had recognized that MVK deficiency could cause inflammatory and CNS disorders60, suggesting that cholesterol biosynthesis in HBMECs may be impacted by PQ exposure. The next step of this cholesterol cascade is the isopentenyl-diphosphate Delta-isomerase 1 (IDI1) that catalyzes isopenty-pyrophosphate into farnesyl-pyrophosphate61. As shown in our results, IDI1 was significantly decreased by PQ exposure in HBMECs. Some studies have demonstrated that cellular expression of IDI1 could be inhibited by ROS generation, resulting in a diminution of lipophilic molecules formation such as ubiquinone, sterols and terpenoids. This observation is suggesting a similar effect induced by ROS generation of the PQ in our study62,63. In addition, a recent study of Tan et al. reported a reduction in cholesterol level, as well as a mitochondrial dysfunction on HBMECs by atorvastatin, which is an inhibitor of cholesterol production64. Our study has also highlighted a significant decrease of cellular cholesterol in a time- and dose-dependent manner. These results are corroborated with the downregulation of key proteins involved in the cholesterol biosynthesis found by DIA quantification and shown by enrichment pathway analysis. A previous study demonstrated the reduction of cholesterol concentration in PQ-treated astrocytes which might support our hypothesis45. These combined observations suggested a PQ-induced toxicity at cellular cholesterol level in brain cells, as astrocytes are tightly in interaction with brain endothelial cells in the BBB. Consequently, dysregulated proteins of the cholesterol cascade and decreased cholesterol level observed in PQ-treated HBMECs could actively affect cell membrane structure and compromise its integrity, by disrupting cellular homeostasis as well as leading to cellular death65. Indeed, the brain demands a constant level of cholesterol in physiological conditions, underlying the need to maintain an optimal cholesterol level for energetic metabolism, cell membrane composition as well as myelination66. As PQ exposure on HBMECs revealed a decreased level of cholesterol, it can be hypothesized that the previously cited processes may be impaired. To add, a causal relationship was reported with oxidative stress generation and reduction of cellular cholesterol in brain diseases67, displaying the potential link between oxidative stress exert by PQ and alteration of the cholesterol metabolism on human brain endothelial cells.
In conclusion, this study underlined that most of the differentially expressed proteins in PQ-treated HBMECs were involved in redox pathways. As expected, mitochondrial dysfunction was denoted after PQ exposure on HBMECs in a dose-dependent manner. Both oxidative stress, demonstrated by affected ubiquinone metabolism and the decrease of mitochondrial maximal respiration, might strongly alter endothelial function by impairing maintenance of the BBB integrity23,24.
This study also highlighted for the first time that PQ is able to modify protein level of proteins involved in cholesterol biosynthesis as well as reduce cellular cholesterol level. Modulation of cholesterol biosynthesis may potentially affect cell membrane structure and compromise its integrity, having a direct effect on energetic metabolism or myelinisation process in brain.
Given the ethical complexity to study a toxicant effect on human population, this in vitro research gives novel insights of PQ toxicity as it pointed out cholesterol alteration on HBMECs. This additional biological alteration is reinforcing the need to definitively ban PQ as an herbicide or to investigate for a safer solution for agricultural workers.
Methods
Cell culture and treatments
Primary human brain microvascular endothelial cells (ACBRI 367, Cell Systems) were cultured onto a rat tail collagen type I-coated (15 µg/mL, Merck Millipore) plates at 50,000 cells/cm2 and maintained in complete endothelial cell growth medium-2 (EGM-2MV BulletKit, Lonza) at 37 °C in a 5% CO2 incubator. At 80% of confluence, cells were treated with Paraquat (Sigma Aldrich) (1, 10 and 100 µM) for 24 h. Afterwards, cells were detached with Stempro Accutase (Gibco) and washed three times with ice-cold Phosphate Buffered Saline (PBS, Gibco) and dry-stored at − 80 °C.
MTS proliferation and LDH cytotoxicity assay
HBMEC were seeded in a 96-well plate (10,000 cells per well) and treated for 24 h with PQ at different concentrations (0.1, 1, 10, 100, 1000 and 5000 µM). Cell proliferation was determined using the MTS assay (CellTiter 96 AQueous One Solution Cell Proliferation Assay, Promega), whereas cytotoxicity was assessed by measuring lactate dehydrogenase (LDH) released using a Pierce LDH cytotoxicity kit (Thermo Scientific). Both the MTS and LDH assays were performed according to the manufacturer’s recommendations.
Sample preparation for mass spectrometry-based proteomics
Cell pellets were resuspended in 0.1% RapiGest (Waters) and 100 mM TEAB (Sigma Aldrich), sonicated (five cycles of 20 s with breaks on ice), and incubated for 10 min at 80 °C. Samples were then spun down (14,000g, 10 min, 4 °C) and the supernatant was recovered. The protein content was measured using Bradford assay (BioRad).
For each sample, 20 μg proteins was reduced using TCEP (final concentration 5 mM, 30 min, 37 °C) (Sigma Aldrich), alkylated using iodoacetamide (final concentration 15 mM, 60 min, RT, in dark condition) (Sigma Aldrich) and digested by an overnight tryptic digestion (w/w ratio 1:50) (Promega). The RapiGest surfactant was cleaved by incubating samples with 0.5% trifluoacetic acid (Sigma Aldrich) (45 min, 37 °C). Samples were then desalted on a C18 reverse phase columns (Harvard Apparatus), peptides were dried under vaccum and subsequently resuspended in 5% ACN 0.1% FA (peptides final concentration of 0.5 μg/μL and spiked with iRT peptide (Biognosys) (1:20)).
MS data independent acquisition and data analysis
For each sample, the equivalent of 2 μg of peptides were analyzed using Liquid Chromatography-Electrospray ionization-MS/MS (LC–ESI–MS/MS) on an Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Fischer Scientific) equipped with an EASY nLC1200 liquid chromatography system (Thermo Fisher Scientific). Peptides were trapped on a 2 cm × 75 μm i.d. PepMap C18 precolumn packed with 3 μm particles and 100 Å pore size. Separation was performed using a 50 cm × 75 μm i.d. PepMap C18 column packed with 2 μm and 100 Å particles and heated at 50 °C. Peptides were separated using a 160 min segmented gradient of 0.1% formic acid (solvent A) and 80% acteonitril 0.1% formic acid (solvent B) (Supplementary Table S4), at a flow rate of 250 nl/min. Data-Independant Acquisition (DIA) was performed with MS1 full scan at a resolution of 60,000 (FWHM) MS1 was acquired in the Orbitrap with an AGC target of 3 × 106, a maximum injection time of 100 ms, a scan range from 400 to 1250 m/z followed by 30 DIA MS2 scan with variable windows. DIA MS2 was performed in the Orbitrap using higher-energy collisional dissociation (HCD) at 30%. AGC target of 2 × 106 and a maximum injection time of 80 ms. The raw DIA MS data were matched against the spectral library following the published protocol68.
Protein abundances were exported from Spectronaut. We also used peptides intensities which were exported and analyzed using mapDIA69. No further normalization was applied. The following parameters were used: min peptides = 1, max peptides = 10, min correl = − 1. Min_DE = 0.01, max_DE = 0.99, and experimental_design = independent design. Proteins were considered to have significantly changed in abundance with a LFDR < 0.05 and an absolute fold change (|FC|) > 1.2.
Biological pathway analysis
The list of differentially expressed proteins from PQ-treated HBMECs (Supplementary Tables S1, S2 and S3) were submitted to MetaCore version 21.2 (Clarivate Analytics) to highlight significantly represented biological pathways. Top 10 biological pathways were selected.
Mitochondrial function—XF cell mito stress test
Mitochondrial respiration was measured using a XF96 extracellular flux analyzer (Seahorse Bioscience, Agilent). The provided 96 well Agilent Seahorse XF Cell Culture Microplate was coated with a solution of rat tail collagen type I (15 µg/mL, Merck Millipore). HBMECs were seeded at a density of 75,000 cells/well, treated with PQ at 1, 10 and 100 µM and maintained in complete endothelial cell growth medium-2 (EGM-2MV BulletKit, Lonza) at 37 °C in a 5% CO2 incubator for 24 h. The sensor cartridge was hydrated with the provided XF Calibrant at 37 °C in a non-CO2 incubator overnight. The culture medium was refreshed 1 h prior to the assay using an optimized medium containing complete endothelial cell growth medium-2 (EGM-2MV BulletKit, Lonza) without serum and with HEPES (final concentration 20 mM, Gibco). Microplate and four mitochondrial inhibitor drugs were subsequently loaded to the hydrated cartridge after reached the optimal concentration for each compound according the manufacturer’s protocol. Briefly, oligomycin (final concentration 4 µM, Sigma-Aldrich), FCCP (final concentration 16 µM, Sigma-Aldrich) and rotenone and antimycin A (final concentration 2 µM, Sigma-Aldrich) were loaded to the hydrated cartridge. All the parameters were considered as explained in Smolina et al. paper70.
Cholesterol assay
HBMECs were seeded in a 96 opaque-walled assay plate (40,000 cells per well) and treated for 24 h with PQ at different concentrations (1, 10, 100 µM). Water was used as control and spiked cholesterol at 60 µM as positive control. Cholesterol was measured using a cholesterol kit assay (Cholesterol Assay, Promega). The cholesterol assay was performed according to the manufacturer’s recommendation.
Statistical analysis
Data are reported as mean ± standard deviation (S.D.). p < 0.05 was considered as statistically significant. Significance is denoted as *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. The data were analyzed using multiple t-test comparisons or one-way analysis of variance (ANOVA).
Data availability
Data are available via ProteomeXchange with identifier PXD026975, username: reviewer_pxd026975@ebi.ac.uk, password: jjlUHMR5.
Abbreviations
- PQ:
-
Paraquat
- ROS:
-
Reactive oxygen species
- HBMECs:
-
Human brain microvascular endothelial cells
- BBB:
-
Blood–brain barrier
- NVU:
-
Neurovascular unit
- DIA-MS:
-
Data-independent acquisition-mass spectrometry
- DIA:
-
Data independent acquisition
- MS:
-
Mass spectrometry
- DEPs:
-
Differentially expressed proteins
- FC:
-
Fold change
- LFDR:
-
Local false discovery rate
References
Ball, N., Teo, W.-P., Chandra, S. & Chapman, J. Parkinson’s disease and the environment. Front. Neurol. 10, 218–218. https://doi.org/10.3389/fneur.2019.00218 (2019).
Brown, T. P., Rumsby, P. C., Capleton, A. C., Rushton, L. & Levy, L. S. Pesticides and Parkinson’s disease—Is there a link?. Environ. Health Perspect. 114, 156–164. https://doi.org/10.1289/ehp.8095 (2006).
Dinis-Oliveira, R. J. et al. Paraquat exposure as an etiological factor of Parkinson’s disease. Neurotoxicology 27, 1110–1122. https://doi.org/10.1016/j.neuro.2006.05.012 (2006).
Goldman, S. M. et al. Genetic modification of the association of paraquat and Parkinson’s disease. Mov. Disord. 27, 1652–1658. https://doi.org/10.1002/mds.25216 (2012).
Tanner, C. M. et al. Rotenone, paraquat, and Parkinson’s disease. Environ. Health Perspect. 119, 866–872. https://doi.org/10.1289/ehp.1002839 (2011).
Shimizu, K. et al. Carrier-mediated processes in blood–brain barrier penetration and neural uptake of paraquat. Brain Res. 906, 135–142. https://doi.org/10.1016/S0006-8993(01)02577-X (2001).
Yuan, G. et al. Simultaneous determination of paraquat and diquat in human plasma by HPLC-DAD: Its application in acute poisoning patients induced by these two herbicides. J. Clin. Lab. Anal. 35, e23669. https://doi.org/10.1002/jcla.23669 (2021).
Baharuddin, M. R., Sahid, I. B., Noor, M. A., Sulaiman, N. & Othman, F. Pesticide risk assessment: A study on inhalation and dermal exposure to 2,4-D and paraquat among Malaysian paddy farmers. J. Environ. Sci. Health B 46, 600–607. https://doi.org/10.1080/03601234.2011.589309 (2011).
Cha, E. S. et al. Paraquat application and respiratory health effects among South Korean farmers. Occup. Environ. Med. 69, 398–403. https://doi.org/10.1136/oemed-2011-100244 (2012).
Thi Hue, N., Nguyen, T. P. M., Nam, H. & Hoang Tung, N. Paraquat in surface water of some streams in Mai Chau Province, the Northern Vietnam: Concentrations, profiles, and human risk assessments. J. Chem. 2018, 8521012. https://doi.org/10.1155/2018/8521012 (2018).
Chatzi, L. et al. Association of allergic rhinitis with pesticide use among grape farmers in Crete, Greece. Occup. Environ. Med. 64, 417–421. https://doi.org/10.1136/oem.2006.029835 (2007).
Du, G. et al. Microstructural changes in the substantia nigra of asymptomatic agricultural workers. Neurotoxicol. Teratol. 41, 60–64. https://doi.org/10.1016/j.ntt.2013.12.001 (2014).
Pezzoli, G. & Cereda, E. Exposure to pesticides or solvents and risk of Parkinson disease. Neurology 80, 2035–2041. https://doi.org/10.1212/WNL.0b013e318294b3c8 (2013).
JUDGMENT OF THE COURT OF FIRST INSTANCE (Second Chamber, Extended Composition) 11 July 2007 (Directive 91/414/EEC – Plant protection products – Paraquat as an active substance – Marketing authorisation – Authorisation procedure – Protection of human and animal health) https://eur-lex.europa.eu/legal-content/EN/TXT/HTML/?uri=CELEX:62004TJ0229&from=GA
Drechsel, D. A. & Patel, M. Chapter 21 Paraquat‐Induced Production of Reactive Oxygen Species in Brain Mitochondria. In Methods in Enzymology Vol. 456, 381–393 (Academic Press, 2009).
Bus, J. S. & Gibson, J. E. Paraquat: model for oxidant-initiated toxicity. Environ. Health Perspect. 55, 37–46. https://doi.org/10.1289/ehp.845537 (1984).
von Tobel, J. S. et al. Immediate and delayed effects of subchronic Paraquat exposure during an early differentiation stage in 3D-rat brain cell cultures. Toxicol. Lett. 230, 188–197. https://doi.org/10.1016/j.toxlet.2014.02.001 (2014).
Castello, P. R., Drechsel, D. A. & Patel, M. Mitochondria are a major source of paraquat-induced reactive oxygen species production in the brain. J. Biol. Chem. 282, 14186–14193. https://doi.org/10.1074/jbc.M700827200 (2007).
Gomez, C., Bandez, M. J. & Navarro, A. Pesticides and impairment of mitochondrial function in relation with the parkinsonian syndrome. Front. Biosci. 12, 1079–1093. https://doi.org/10.2741/2128 (2007).
Smeyne, R. J. et al. Assessment of the effects of MPTP and paraquat on dopaminergic neurons and microglia in the substantia nigra pars compacta of C57BL/6 mice. PLoS ONE 11, e0164094. https://doi.org/10.1371/journal.pone.0164094 (2016).
Serlin, Y., Shelef, I., Knyazer, B. & Friedman, A. Anatomy and physiology of the blood-brain barrier. Semin. Cell Dev. Biol. 38, 2–6. https://doi.org/10.1016/j.semcdb.2015.01.002 (2015).
Knowland, D. et al. Stepwise recruitment of transcellular and paracellular pathways underlies blood-brain barrier breakdown in stroke. Neuron 82, 603–617. https://doi.org/10.1016/j.neuron.2014.03.003 (2014).
Sweeney, M. D., Zhao, Z., Montagne, A., Nelson, A. R. & Zlokovic, B. V. Blood–brain barrier: From physiology to disease and back. Physiol. Rev. 99, 21–78. https://doi.org/10.1152/physrev.00050.2017 (2019).
Bernacki, J., Dobrowolska, A., Nierwińska, K. & Małecki, A. Physiology and pharmacological role of the blood–brain barrier. Pharmacol. Rep. 60, 600–622 (2008).
Saidijam, M., Karimi Dermani, F., Sohrabi, S. & Patching, S. G. Efflux proteins at the blood–brain barrier: Review and bioinformatics analysis. Xenobiotica Fate Foreign Comp. Biol. Syst. 48, 506–532. https://doi.org/10.1080/00498254.2017.1328148 (2018).
Barar, J., Rafi, M. A., Pourseif, M. M. & Omidi, Y. Blood-brain barrier transport machineries and targeted therapy of brain diseases. Bioimpacts 6, 225–248. https://doi.org/10.15171/bi.2016.30 (2016).
Ramos-Cabrer, P. & Campos, F. Liposomes and nanotechnology in drug development: focus on neurological targets. Int. J. Nanomed. 8, 951–960. https://doi.org/10.2147/IJN.S30721 (2013).
Sweeney, M. D., Sagare, A. P. & Zlokovic, B. V. Blood–brain barrier breakdown in Alzheimer disease and other neurodegenerative disorders. Nat. Rev. Neurol. 14, 133. https://doi.org/10.1038/nrneurol.2017.188 (2018).
Rochfort, K. D., Collins, L. E., Murphy, R. P. & Cummins, P. M. Downregulation of blood-brain barrier phenotype by proinflammatory cytokines involves NADPH oxidase-dependent ROS generation: consequences for interendothelial adherens and tight junctions. PLoS ONE 9, e101815. https://doi.org/10.1371/journal.pone.0101815 (2014).
Kuriakose, M. et al. Synergistic role of oxidative stress and blood–brain barrier permeability as injury mechanisms in the acute pathophysiology of blast-induced neurotrauma. Sci. Rep. 9, 7717. https://doi.org/10.1038/s41598-019-44147-w (2019).
Rathinam, M. L. et al. Astrocyte mediated protection of fetal cerebral cortical neurons from rotenone and paraquat. Environ. Toxicol. Pharmacol. 33, 353–360. https://doi.org/10.1016/j.etap.2011.12.027 (2012).
Yang, W. & Sun, A. Y. Paraquat-induced free radical reaction in mouse brain microsomes. Neurochem. Res. 23, 47–53. https://doi.org/10.1023/a:1022497319548 (1998).
Wang, X., Luo, F. & Zhao, H. Paraquat-Induced reactive oxygen species inhibit neutrophil apoptosis via a p38 MAPK/NF-κB–IL-6/TNF-α positive-feedback circuit. PLoS ONE 9, e93837. https://doi.org/10.1371/journal.pone.0093837 (2014).
Tawara, T. et al. Effects of paraquat on mitochondrial electron transport system and catecholamine contents in rat brain. Arch. Toxicol. 70, 585–589. https://doi.org/10.1007/s002040050316 (1996).
Sharma, L. K., Lu, J. & Bai, Y. Mitochondrial respiratory complex I: Structure, function and implication in human diseases. Curr. Med. Chem. 16, 1266–1277. https://doi.org/10.2174/092986709787846578 (2009).
Gear, A. J. L., Ahrenholz, D. H. & Solem, L. D. Paraquat poisoning in a burn patient. J. Burn Care Rehabil. 22, 347–351. https://doi.org/10.1097/00004630-200109000-00011 (2001).
Hernández, A. F. et al. Low level of exposure to pesticides leads to lung dysfunction in occupationally exposed subjects. Inhal. Toxicol. 20, 839–849. https://doi.org/10.1080/08958370801905524 (2008).
Berry, C., La Vecchia, C. & Nicotera, P. Paraquat and Parkinson’s disease. Cell Death Differ. 17, 1115–1125. https://doi.org/10.1038/cdd.2009.217 (2010).
Freire, C. & Koifman, S. Pesticide exposure and Parkinson’s disease: Epidemiological evidence of association. Neurotoxicology 33, 947–971. https://doi.org/10.1016/j.neuro.2012.05.011 (2012).
Bus, J. S., Aust, S. D. & Gibson, J. E. Paraquat toxicity: Proposed mechanism of action involving lipid peroxidation. Environ. Health Perspect. 16, 139–146. https://doi.org/10.1289/ehp.7616139 (1976).
Sandström, J. et al. Potential mechanisms of development-dependent adverse effects of the herbicide paraquat in 3D rat brain cell cultures. Neurotoxicology 60, 116–124. https://doi.org/10.1016/j.neuro.2017.04.010 (2017).
McCarthy, S., Somayajulu, M., Sikorska, M., Borowy-Borowski, H. & Pandey, S. Paraquat induces oxidative stress and neuronal cell death; neuroprotection by water-soluble Coenzyme Q10. Toxicol. Appl. Pharmacol. 201, 21–31. https://doi.org/10.1016/j.taap.2004.04.019 (2004).
Bagley, A. C., Krall, J. & Lynch, R. E. Superoxide mediates the toxicity of paraquat for Chinese hamster ovary cells. Proc. Natl. Acad. Sci. USA 83, 3189–3193. https://doi.org/10.1073/pnas.83.10.3189 (1986).
Yang, Y. et al. Inflammation leads to distinct populations of extracellular vesicles from microglia. J. Neuroinflam. 15, 168. https://doi.org/10.1186/s12974-018-1204-7 (2018).
Li, Z., Zheng, J. & Zhang, X.-F. Detrimental effects of paraquat on astrocytes-regulating synaptic functions. Dose–Response 16, 1559325818761681. https://doi.org/10.1177/1559325818761681 (2018).
Fukushima, T. et al. Mechanism of cytotoxicity of paraquat: III. The effects of acute paraquat exposure on the electron transport system in rat mitochondria. Exp. Toxicol. Pathol. 46, 437–441. https://doi.org/10.1016/S0940-2993(11)80056-4 (1994).
Richardson, J. R., Quan, Y., Sherer, T. B., Greenamyre, J. T. & Miller, G. W. Paraquat neurotoxicity is distinct from that of MPTP and rotenone. Toxicol. Sci. 88, 193–201. https://doi.org/10.1093/toxsci/kfi304 (2005).
Yang, W. & Tiffany-Castiglioni, E. The bipyridyl herbicide paraquat induces proteasome dysfunction in human neuroblastoma SH-SY5Y cells. J. Toxicol. Environ. Health A 70, 1849–1857. https://doi.org/10.1080/15287390701459262 (2007).
Yang, W. & Tiffany-Castiglioni, E. Paraquat-induced apoptosis in human neuroblastoma SH-SY5Y cells: involvement of p53 and mitochondria. J. Toxicol. Environ. Health A 71, 289–299. https://doi.org/10.1080/15287390701738467 (2008).
Li, Z., Zheng, J. & Zhang, X.-F. Detrimental effects of paraquat on astrocytes-regulating synaptic functions. Dose Response 16, 1559325818761681–1559325818761681. https://doi.org/10.1177/1559325818761681 (2018).
Bergman, O. & Ben-Shachar, D. Mitochondrial oxidative phosphorylation system (OXPHOS) deficits in schizophrenia: Possible interactions with cellular processes. Can. J. Psychiatry 61, 457–469. https://doi.org/10.1177/0706743716648290 (2016).
Goldstein, J. L. & Brown, M. S. Regulation of the mevalonate pathway. Nature 343, 425–430. https://doi.org/10.1038/343425a0 (1990).
Caballero, B. Cholesterol. In Encyclopedia of Human Nutrition (2nd Edition) 379–385 (Elsevier, 2005).
Ridgway, N. & McLeod, R. Cholesterol synthesis. In Biochemistry of Lipids, Lipoproteins and Membranes (6th Edition) 327–358 (Elsevier, 2016).
Dietschy, J. M. & Turley, S. D. Cholesterol metabolism in the brain. Curr. Opin. Lipidol. 12, 105–112. https://doi.org/10.1097/00041433-200104000-00003 (2001).
Vance, J. E. Dysregulation of cholesterol balance in the brain: Contribution to neurodegenerative diseases. Dis. Model. Mech. 5, 746–755. https://doi.org/10.1242/dmm.010124 (2012).
Rokosz, L. L. et al. Human cytoplasmic 3-hydroxy-3-methylglutaryl coenzyme a synthase: Expression, purification, and characterization of recombinant wild-type and Cys129 Mutant enzymes. Arch. Biochem. Biophys. 312, 1–13. https://doi.org/10.1006/abbi.1994.1273 (1994).
Mullen, P. J., Yu, R., Longo, J., Archer, M. C. & Penn, L. Z. The interplay between cell signalling and the mevalonate pathway in cancer. Nat. Rev. Cancer 16, 718–731. https://doi.org/10.1038/nrc.2016.76 (2016).
Smith, S. C., Robinson, A. R., Niedernhofer, L. J. & Hetman, M. Downregulation of cholesterol biosynthesis genes in the forebrain of ERCC1-deficient mice. Neurobiol. Dis. 45, 1136–1144. https://doi.org/10.1016/j.nbd.2011.12.036 (2012).
Favier, L. A. & Schulert, G. S. Mevalonate kinase deficiency: Current perspectives. Appl. Clin. Genet. 9, 101–110. https://doi.org/10.2147/TACG.S93933 (2016).
Hahn, F. M., Xuan, J. W., Chambers, A. F. & Poulter, C. D. Human isopentenyl diphosphate: Dimethylallyl diphosphate isomerase: Overproduction, purification, and characterization. Arch. Biochem. Biophys. 332, 30–34. https://doi.org/10.1006/abbi.1996.0312 (1996).
Fang, C.-Y. et al. Idi1 and Hmgcs2 are affected by stretch in HL-1 Atrial myocytes. Int. J. Mol. Sci. 19, 4094. https://doi.org/10.3390/ijms19124094 (2018).
Chen, Y.-W. et al. Cardioprotective effects of quercetin in cardiomyocyte under ischemia/reperfusion injury. Evid.-Based Complement. Alternative Med. 2013, 364519. https://doi.org/10.1155/2013/364519 (2013).
Tan, Q., Yu, D. & Song, L. Atorvastatin disrupts primary human brain microvascular endothelial cell functions via prenylation-dependent mitochondrial inhibition and oxidative stress. Fundam. Clin. Pharmacol. https://doi.org/10.1111/fcp.12615 (2020).
Ammendolia, D. A., Bement, W. M. & Brumell, J. H. Plasma membrane integrity: Implications for health and disease. BMC Biol. 19, 71. https://doi.org/10.1186/s12915-021-00972-y (2021).
Czuba, E., Steliga, A., Lietzau, G. & Kowiański, P. Cholesterol as a modifying agent of the neurovascular unit structure and function under physiological and pathological conditions. Metab. Brain Dis. 32, 935–948. https://doi.org/10.1007/s11011-017-0015-3 (2017).
Martín, M. G., Pfrieger, F. & Dotti, C. G. Cholesterol in brain disease: Sometimes determinant and frequently implicated. EMBO Rep. 15, 1036–1052. https://doi.org/10.15252/embr.201439225 (2014).
Dozio, V. & Sanchez, J. C. Profiling the proteomic inflammatory state of human astrocytes using DIA mass spectrometry. J. Neuroinflam. 15, 331. https://doi.org/10.1186/s12974-018-1371-6 (2018).
Teo, G. et al. mapDIA: Preprocessing and statistical analysis of quantitative proteomics data from data independent acquisition mass spectrometry. J. Proteom. 129, 108–120. https://doi.org/10.1016/j.jprot.2015.09.013 (2015).
Gilbert, D. F., & Friedrich, O. Assaying Mitochondrial Respiration as an Indicator of Cellular Metabolism and Fitness. In Cell Viability Assays: Methods and Protocols 79–87 (Springer, New York, 2017).
Acknowledgements
We are grateful to the Swiss Centre for Applied Human Toxicology for funding this study. We also want to thank Alexandre Hainard, Carla Pasquarello and Patrizia Arboit from the proteomic core facility of the Geneva University for their kind help and advice for the proteomic analyses by mass spectrometry. A special thanks is required to the R.E.A.D.S UNIT at the University of Geneva, in particular Yves Cambet, for his valuable help and availability.
Author information
Authors and Affiliations
Contributions
T.V., D.S. and J.S.C., designed the study, provided the discussion and suggestions to the experiments. D.S. helped visualization, conceptualization and curation of data. J.C.S. project administration and funding acquisition. T.V. performed the experiments, analyzed and interpreted data, acquired data and wrote the original manuscript with input from all authors. All authors read and approved to the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Tatjana, V., Domitille, S. & Jean-Charles, S. Paraquat-induced cholesterol biosynthesis proteins dysregulation in human brain microvascular endothelial cells. Sci Rep 11, 18137 (2021). https://doi.org/10.1038/s41598-021-97175-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-97175-w
This article is cited by
-
SeFilter-DIA: Squeeze-and-Excitation Network for Filtering High-Confidence Peptides of Data-Independent Acquisition Proteomics
Interdisciplinary Sciences: Computational Life Sciences (2024)
-
Environmental Chemical Exposures and Mitochondrial Dysfunction: a Review of Recent Literature
Current Environmental Health Reports (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.