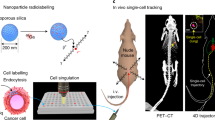
Abstract
Despite the tremendous advancements made in cell tracking, in vivo imaging and volumetric analysis, it remains difficult to accurately quantify the number of infused cells following stem cell therapy, especially at the single cell level, mainly due to the sensitivity of cells. In this study, we demonstrate the utility of both liquid scintillator counter (LSC) and accelerator mass spectrometry (AMS) in investigating the distribution and quantification of radioisotope labeled adipocyte derived mesenchymal stem cells (AD-MSCs) at the single cell level after intravenous (IV) transplantation. We first show the incorporation of 14C-thymidine (5 nCi/ml, 24.2 ng/ml) into AD-MSCs without affecting key biological characteristics. These cells were then utilized to track and quantify the distribution of AD-MSCs delivered through the tail vein by AMS, revealing the number of AD-MSCs existing within different organs per mg and per organ at different time points. Notably, the results show that this highly sensitive approach can quantify one cell per mg which effectively means that AD-MSCs can be detected in various tissues at the single cell level. While the significance of these cells is yet to be elucidated, we show that it is possible to accurately depict the pattern of distribution and quantify AD-MSCs in living tissue. This approach can serve to incrementally build profiles of biodistribution for stem cells such as MSCs which is essential for both research and therapeutic purposes.
Similar content being viewed by others
Introduction
The ability to label, track, and image stem cells in real time following transplantation has enabled a wide range of in depth studies that aim to explore and comprehend important biological phenomenon pertinent to regenerative medicine and in particular, stem cell therapy. The use of various types of nanoparticles including quantum dots, superparamagnetic iron oxides (SPIO), silica, and gold in conjunction with fluorescent and photoacoustic imaging techniques as well as other high resolution in vivo imaging methods such as positron emission tomography (PET) and single-photon emission tomography (SPECT), magnetic resonance imaging (MRI), and X-Ray computed microtomography (microCT) has allowed for the long term monitoring of stem cells in model organisms to better understand the behavior of therapeutic cells in both natural and diseased environments1,2,3,4,5,6,7,8.
Despite the tremendous advancements made in cell tracking, it remains quite difficult to accurately quantify the number of infused cells within living tissues, especially at the single cell level, mainly due to the sensitivity of cells. And while quantitative PCR is widely used to detect, characterize, and quantify nucleic acids, the method is only relative in the sense that the results will always be proportional to the amount of replicated DNA as opposed to the actual number of active cells9,10. Therefore, radioisotope labeling such as carbon-14 thymidine (14C-thymidine) which incorporates into new DNA, paired with a liquid scintillator counter (LSC) is commonly employed to measure the radioactivity of 14C labeled cells for long-term tracking and quantitative analysis11,12. A previous study has shown the tracking of placenta-derived mesenchymal stem cells (PDB-MSCs) in nude mice via 14C labeling after IV transplantation, concluding that it is a stable, long term, and quantitative cell tracker12. However, LSC is incapable of measuring trace amounts of radioactivity due to its low sensitivity and higher radioisotope concentrations can often affect the functional potential of stem cells such as engraftment, proliferation, secretion, and differentiation. Therefore, a highly sensitive (limits of detection are indicated in terms of attmole (10–18) ~ zeptomole (10–21) quantities of 14C in small samples) method such as accelerator mass spectrometry (AMS) is required to quantify stem cells labeled with low 14C thymidine radioactivity without interfering with their normal function13. Originally developed for the field of radiocarbon dating in the late 1970s, AMS has proven to be an effective instrument for biomedical and pharmaceutical research when trace quantities of 14C need to be quantified in small samples14,15. Recently, the tracking of tumor colonization was demonstrated by AMS in a xenograft model which served to quantitatively evaluate metastasis and tumor aggressiveness16. While the biological mechanisms and significance behind the distribution of cells throughout time remains largely a mystery due to the limited number of studies, a wider array of data could help to identify general patterns in distribution and localization dependent on cell type and route of delivery.
Herein, we demonstrate using AMS in investigating the distribution and localization of adipocyte derived mesenchymal stem cells (AD-MSCs) at the single cell level after intravenous (IV) transplantation. This study corroborates some of the findings made in previous work but it also reveals the localization of single cells in unexpected organs at early time points. While the significance of these cells is yet to be elucidated, we show that it is possible to accurately depict the pattern of distribution and localization of AD-MSCs in living tissues over time.
Results
Selection of optimal concentration for 14 C-labeling AD-MSCs
Since 14C-thymidine incorporates into the newly replicated DNA when the cell divides, we simply rationalized that AD-MSCs exhibiting a higher proliferation rate would incorporate higher amounts of 14C-thymidine during cultivation. Therefore, 2.5 nCi/ml of 14C-thymidine was added into various medium conditions (10% FBS, 20% FBS, 10% FBS + bFGF, and 20% FBS + bFGF) to determine the optimal culture group for cell proliferation and consequently, 14C-thymidine incorporation (Fig. 1A). Cell counting was performed on day 6 which revealed the proliferative capacity of each group ranking from highest to lowest; 20% FBS + bFGF > 20% FBS > 10% FBS + bFGF > 10% FBS (Fig. 1B). Interestingly and contrary to our expectation, 14C activity was significantly higher in the groups containing 10% FBS, especially when supplemented with bFGF, despite the higher proliferation rate observed in those containing 20% FBS (Fig. 1C).
Optimization of 14C thymidine concentration for AD-MSCs. (A) Method of labeling to prepare 14C-labeled AD-MSCs. (B) Comparison of cell number by different culture medium condition. (C) Comparison of 14C activity by different culture medium condition. (D) Image for identifying cell number and morphology change by different 14C thymidine concentration. (E) Cell number comparison by different 14C thymidine concentration using hemocytometer. (F) Comparison of cell proliferation by different 14C thymidine concentration using CCK-8. (G) Comparison of uptake rate of AD-MSCs labeled with different concentration. (H) Measurement of radioactivity of DNA labeled with different 14C thymidine concentration. (I) Comparison of extraction rate of DNA. ***p < 0.001 relative to the control or 5 nCi/ml, &&&p < 0.001 relative to the 10% FBS + bFGF group, ###p < 0.001 relative to the 20% FBS group. All data are expressed as mean ± SD.
Thus, the 10% FBS + bFGF group was selected to be best suited for both AD-MSC proliferation as well as 14C-thymidine incorporation. Next, we sought to determine whether increasing concentrations of 14C-thymidine would negatively impact AD-MSC proliferation and morphology by comparing cultivations that received 0 (control), 1, 2.5, 5, 10, or 25 nCi/ml of 14C-thymidine (Supplementary Table S1). Based on observation at day 6, it was clear that concentrations of 10 nCi/ml and 25 nCi/ml reduced the proliferation rate of AD-MSCs as more vacant spaces were readily identified in both groups (Fig. 1D). This was further confirmed by cell counting (hemocytometer) and CCK-8 assay which showed a marked decrease in both the total and viable cell population, respectively (Fig. 1E,F). When the level of 14C-thymidine uptake for each group was measured as a percentage by LSC, it revealed a decline in uptake efficiency for AD-MSCs that received concentrations of 10 and 25 nCi/ml (Fig. 1G). As expected, higher levels of 14C activity was detected as the concentration increased (Fig. 1H) and the 14C-thymidine incorporated DNA was able to be extracted for all groups (Fig. 1I). Based on these results, a concentration of 5 nCi/ml was determined to be the optimal concentration for AD-MSCs because 14C activity was higher than the 1 and 2.5 nCi/ml groups while the cell proliferation rate and uptake efficiency was maintained, unlike the 10 and 25 nCi/ml groups which exhibited adverse effects.
Characterization of 14C-labeled AD-MSCs
Unlabeled and 14C-labeled AD-MSCs were characterized by specific CD marker expression and multi-lineage differentiation to compare surface characters and functional potential. FACS analysis revealed that both populations displayed the hallmark MSC antigen profile of positive CD73, CD90, and CD105 complemented by negative CD34 and CD45 (Fig. 2A). However, a noticeable decrease in CD105 expression was detected as 92.72% of the unlabeled AD-MSCs expressed CD105 which was higher than the 80.62% expressed by the labeled cells. While it remains unclear as to why a difference in marker expression emerged.
Analysis of biological characteristics of 14C-labeled AD-MSCs. (A) Confirmation of MSC specific marker expression of unlabeled and 14C-labeled AD-MSCs by FACS analysis. (B) Investigation of multipotent properties (adipogenesis, osteogenesis, chondrogenesis) of unlabeled and 14C-labeled AD-MSCs. (C) Comparison of cell cycle analysis of unlabeled and 14C-labeled AD-MSCs. Scale bar = 200 µm.
The functional characteristics of AD-MSCs and its multipotency were investigated through differentiation. Both groups were subjected to differentiation conditions for adipocytes, osteoblasts, and chondrocytes which were identified by Oil Red O, Alizarin red S, and Alcian blue staining, respectively. Both groups were capable of differentiating into all three cell types, confirming that multipotency was retained in the labeled group (Fig. 2B). In addition, cell cycle analysis was performed to further compare both populations in relation to the different phases of the cell cycle. Both groups shared a very similar profile for the sub-G1, G1, S, and G2-M phases, indicating that cell growth, genome replication, and ultimately the process of mitosis remained unchanged after 14C-labeling (Fig. 2C). These results demonstrated that the key characteristics of AD-MSCs in regards to surface markers, multipotency, and cell division were retained in the labeled cells, suggesting that the incorporation of 14C-thymidine does not affect the key characteristics of AD-MSCs.
Pre-treatment process and calibration the LSC and AMS methods
The detection and quantification of radioactivity by LSC and AMS were compared by first transplanting 50,000 14C-labeled AD-MSCs into a homogenized liver sample before analysis. For LSC, the organ samples were simply solubilized (SOLVABLE, PerkinElmer, Waltham, MA, USA) then decolorized using H2O2 before sample analysis (Fig. 3A). The LSC calibration curve was prepared using 14C activity values (disintegration per minute, dpm) according to the number of labeled AD-MSCs (200, 500, 1000, 2500, 5000, 10,000, and 50,000 cells) and the R2 value was 0.9982, indicating linearity (Fig. 3B). Furthermore, to generate a calibration curve for the AMS analysis, 50,000 labeled AD-MSCs were spiked into the homogenized liver sample and diluted to the indicated number of cells (1, 5, 10, 25, 50, and 100 cells). In contrast to LSC, AMS required a comparatively lengthy process that involves the graphitization of organ samples before analysis which is detailed in the Materials and Methods section (Fig. 3C). The diluted 14C-labeled AD-MSCs were measured by AMS after pretreatment. The calibration curve of the AMS was prepared using modern carbon (MC, the ratio of 14C/12C in the reference atmosphere of the 1950s), and a dynamic range of the AMS from 0.1 MC to 150 MC (0.001356 — 2.034 dpm/mgC) according to the number of labeled AD-MSCs. The R2 value was 0.9997, indicating linearity (Fig. 3D). When comparing the number of detectable AD-MSCs for each assay, it showed that LSC could not detect less than 200 AD-MSCs. However, a value of 3.4 MC in one cell was obtained from AMS analysis, which was 30 times higher than the limit of detection. These results show that LSC is inadequate in detecting a small number of cells and AMS is required for tracking single cell populations.
Pretreatment procedure and quantitative curves of LSC and AMS analysis. (A) Pretreatment procedure for LSC measurement. (B) Calibration curve of LSC using 14C activity value (dpm) according to cell number (200, 500, 1,000, 2,500, 5,000, 10,000, and 50,000 cells) (C) Pretreatment procedure for AMS measurement. (D) Calibration curve of AMS using modern carbon (MC) according to cell number (1, 5, 10, 25, 50, and 100 cells). All data are expressed as mean ± SD.
Quantification of 14C-labeled AD-MSCs in nude mice by LSC and AMS analyses
In order to investigate the distribution and localization of transplanted AD-MSCs in vivo, nude mice were subjected to IV injection (1 \(\times\) 106 cells) and 14C radioactivity was measured in the lung, spleen, liver, heart, kidney, and brain by both LSC and AMS at 4 h, 12 h, 24 h, 48 h, and day 7. Using the calibration curve obtained from Fig. 3, the cell concentration and cell amount were calculated from the LSC and AMS measurement of the sample as shown in the following equation. The linear regression equation of calibration curve for quantitation of cell number using LSC was obtained using the cell number and radioactivity, Eq. (1). The cell number was calculated Eq. (2), (3). Where, Lsample and Lblank are the radioactivity (dpm) of the sample and pre-dose organ, respectively
The measurement results of AMS were obtained by the ratio of the amount of 14C (14Ccell and 14Corgan) and 12C (12Corgan) as shown in the following equation Eq. (4). Rmesu and Rblank are the value of 14C/12C of the sample and pre-dose organ, respectively. 12Corgan is the amounts of carbon (12C) contained in the prepared organ. Rsample was calculated by subtracting Rmesu from Rblank, Eq. (5).
The linear regression equation of calibration curve for quantitation of cell number using AMS was obtained using the cell number and modern carbon, Eq. (6). The cell number was calculated Eqs. (7) and (8).
Cell concentration (cell number/mg) and cell amount (cell number/organ) can be obtained from cell number, the weight of the sample (Wsample), and the weight of the organ (Worgan). Therefore cell concentration and cell amount were calculated, Eq. (9), (10).
For LSC, AD-MSCs were only detected in the lung with the highest number of cells existing within the organ at 4 h post-transplantation (5,188 ± 365 infused cells/mg). This sharply decreased over time (12 h: 2,930 ± 453, 24 h: 1,673 ± 70, 48 h: 1,161 ± 336) with little to no trace left by 72 h (Fig. 4A). In contrast, AMS was able to detect a small number of cells located in all other tested organs but difficult to quantify larger numbers such as the lung’s case due to over-detection. At 4 h post-transplantation, the number of AD-MSCs per mg located in the spleen and liver was 19 ± 8 and 14 ± 1, respectively. And even lesser amounts or single cells were detected in the heart and kidney (1 ± 0 infused cells number/mg) while no cells were detected in the brain. Interestingly, the number of cells increased at 12 h with exception to the lung which may indicate the occurrence of cell migration from highly populated organs such as the lung to other organs such as the liver and spleen. However, the number of cells was reduced in the liver after 24 h as a fourfold, while other organs (spleen, heart, and kidney) were observed after 12 h as a twofold reduction. Even though the decline continued at 48 h, a small number of residual cells remained within each tissue at day 7 (Fig. 4B). Based on the compiled data of both LSC and AMS, we were able to determine the number of infused cells per organ based on the average mass of each organ to better illustrate AD-MSC localization following IV transplantation (Fig. 4C). The majority of AD-MSCs were present in the lung from 4 h (619,266 ± 36,239) to 48 h (157,448 ± 40,908) and even though the number of active cells continued to decline, ~ 5000 ± AD-MSCs lingered within the organ on day 7. As for the liver, the number of cells significantly increased from 4 h (14,796 ± 1,552) to 12 h (21,941 ± 1,060) before a sharp decline at 24 h (4,761 ± 907). These values were interestingly similar to that of the lung on day 7. The spleen also exhibited a rise in population from 4 h (1,497 ± 590) to 12 h (1,993 ± 454) followed by a decline at 24 h (1,018 ± 156) which plateaued at 48 h (884 ± 593) to day 7 (908 ± 674). This pattern of distribution was similar in the heart and kidney at a much lesser extent (Supplementary Table S2).
Quntification and distribution results of infused 14C-labeled AD-MSCs in nude mice using LSC and AMS analysis. (A) The distribution of 14C-labeled AD-MSCs after IV injection in nude mice. 14C-radioactivity of each organ using LSC. Data were represented as mean cells number/mg. (B) Modern carbon (MC) measurement of each organ using AMS. Data were represented as mean cells number/mg. (C) Total amount of cells in organs using 14C radioactivity value from LSC and AMS analysis. All data are expressed as mean ± SD.
Detection of DiI-labeled AD-MSCs in nude mice
To further confirm the successful engraftment of transplanted AD-MSCs and validate the bio-distribution of cells reflected in LSC and AMS, Dil stained AD-MSCs were identified in 5 µm cryosections of each organ at 4 h, 24 h, and 168 h (day 7). And similar to that of 14C-labeled AD-MSCs, we found that the majority of cells populated the lung at 4 h followed by the liver and spleen. We also found that the cells began to clear within the first day yet a small number of residual cells remained within the specified organs on day 7. These results complements the tracking of AMS by showing that cells are engrafted visually, although accurate quantification is not possible within each organ (Fig. 5).
Discussion
This study demonstrates the successful incorporation of 14C-thymidine (5 nCi/ml) into AD-MSCs without affecting key biological characteristics pertinent to morphology, CD marker expression, differentiation, and cell cycle. These cells were then utilized to track and quantify the distribution of AD-MSCs delivered through an intravenous route (tail vein) by AMS, revealing the number of AD-MSCs existing within different organs per mg and per organ at different time points. Notably, the results show that this highly sensitive approach can quantify one cell per mg which effectively means that AD-MSCs can be detected in various tissues at the single cell level.
Even though the history of AMS is extensive such as Geoscience, Archaeology, Environmental Science, and Biomedical Science, its adaptation to the field of regenerative medicine is fairly recent as only a few studies documented its usage in the tracking and quantification of distributed stem cells in vivo. Previously reported radioisotope labeling methods such as 51Cr, 124I, and 99mTC often require highly radioactivity for imaging purposes; 18.5 MBq (500 µCi) for 51Cr, 3.7 MBq/0.1 ml (100 µCi/0.1 ml) for 124I-FIAU, and 3.7 MBq (100 µCi) for 64Cu-PISM17,18. Furthermore, studies that employ a β-counter, often used concentrations of 1 µCi of 3H-thymidine or 14C-thymidine which has also been estimated to cause cell damage12,19,20,21. In this study, we show that far lower concentrations can be used for the long term tracking of AD-MSCs following transplantation due to long half-life of 14C (half-life = 5,370 years). Since stem cells exhibit plasticity unlike somatic cells, it is very likely that they are more prone to change upon interference thereby a highly sensitive approach is advantageous to ensure that its character is not disrupted before administration.
Another important factor for cell distribution is the route of delivery. Although our study is limited to IV, it is well known that other delivery routes, whether systematic (IV) or local (direct injection) can change the distributive outcome. For example in IV and intra-atrial (IA), cells are able to be distributed throughout several organs with most of the cells accumulating within the lung, liver, and spleen as shown in this study which is also consistent with previous reports9,22,23. In contrast, direct injection (DI) can target a specific tissue but prohibits interactions between MSCs and secondary signaling systems24. And because there is no consensus on the optimal delivery route for MSCs at this time, the approach used in this study can help to elucidate the distribution and localization of stem cells following transplantation through a selected route in order to further determine effective routes for various cell types. In sum, radioisotope labeling paired with AMS can be a very useful tool in tracking and quantifying AD-MSCs at the single cell level after transplantation. Future studies should focus on applying a similar approach to other stem cell variants of both multipotent and pluripotent origin to incrementally build profiles of biodistribution which can serve to accelerate our understanding of stem cell behavior in vivo especially in light of the increasing attention and clinical studies surrounding stem cell therapeutics.
Materials and methods
Cell culture and labeling of 14C-tymidine
Human adipose-derived mesenchymal stem cells (hAD-MSCs; ATCC, Manassas, VA, USA) were cultured in Dulbecco’s Modified Eagle Media (DMEM; Gibco BRL, Grand Island, NY) supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Waltham, MA, USA), 1% GlutaMax (Gibco), 1% Non-Essential Amion Acids Solution (MEM NEAA; Gibco), 1% penicillin–streptomycin (P/S; Thermo Fisher Scientific) and 10 ng/ml basic fibroblast growth factor (bFGF). Cells were maintained at 37 °C in a humidified atmosphere containing 5% CO2. For labeling of [2-14C]-thymidine (Moravek Biochemicals, Brea, CA, USA) in the AD-MSC, AD-MSCs were seeded at 7.8 \(\times\) 105 cells in 100 mm culture dish (46 cells/mm2) with culture medium containing 14C-tymidine at the indicated concentration (1, 2.5, 5, 10, and 25 nCi/ml; 4.8, 12.1, 24.2, 48.4, and 121.1 ng/ml). After 2 days and 4 days, the medium was replaced with culture medium containing 14C-tymidine and cells were detached using 0.25% trypsin–EDTA (Gibco) in 6 days.
Cell proliferation assay
Cell viability was examined via a Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Kumamoto, Japan) according to the manufacture’s protocol. Cells were seeded into 96-well plates at a density of 2,000 cells/well in a final volume of 100 µl and grown under culture medium containing indicated concentration of 14C-thymidine. The cells were cultured for 2 day and 6 day; then, 10 µl CCK-8 reagent was added to each well and the cells were incubated at 37 °C for 4 h. Cell viability was calculated by measuring the absorbance at 450 nm with a microplate reader.
DNA extraction
DNA was harvested from 14C labeled AD-MSCs grown in culture. Cultured 14C AD-MSCs were washed in PBS prior to 2 ~ 3 \(\times\) 105 cells was digested with Proteinase K, RNase, and AL buffer using commercially available silica-based columns (DNeasy Blood & Tissue Kit, Qiagen, Hilden, Germany). Concentration of isolated DNA was quantified using a NanoDrop one Microvolume UV–Vis Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA).
Analysis of liquid scintillation counter (LSC)
The activity of the incorporated 14C-thymidine into the DNA of AD-MSCs was measured using Tri-Carb 4910TR LSC instrument (Perkin Elmer, Shelton, CT, USA). 14C labeled AD-MSC and extracted DNA in PBS (DNA concentration, 100 μl) of aliquot from each sample were added into 10 ml of LSC cocktail (Perkin Elmer, Shelton, CT, USA). Each radioactivity in mixed solution was measured through LSC for 30 min.
Characterization of 14C-labeled adipose-derived mesenchymal stem cells
Characterization of AD-MSC was conducted according to our previously described method25. Cells were detached by 0.25% trypsin–EDTA at ~ 90% passage, detached cells were resuspended in FACS buffer (PBS solution including 0.5% bovine albumin (BSA) and 2 mm EDTA) and filtered using a premoistened a 40-µm cell strainer. Cells were then labeled using each antibody of MSC surface markers according to manufacturer’s instructions. Types of antibody are as follows. Fluorochrome-conjugated antibodies for CD73-PE, CD90-APC, and CD105-PE (BD Biosciences, Bedford, MA, USA), along with a negative marker CD34 and CD45 conjugated to PE (BD Biosciences) were used. Also, corresponding IgG controls were prepared equally, and 30,000 labeled cells were acquired and analyzed using Becton Dickinson FACS Calibur (BD Biosciences).
Assessment of multilineage differentiation
Multilineage differentiation of AD-MSC was conducted according to our previously described method25. For the induction of osteoblasts, adipocytes, and chondroblasts, commercially available kits were used (Thermo Fisher Scientific, Waltham, MA, USA). Cells under differentiation conditions were maintained in 4-well plates or 12-well plates. Osteogenesis was incubated for 21 days, and adipogenic and chondrogenic lineage was induced for 14 days. All experimental procedures were performed according to the manufacturer’s instructions. To evaluate each differentiation process, appropriate staining was performed: Alizarin Red S to identify calcium deposits, Oil Red O to detect intracellular lipid droplets, and Alcian blue to confirm the formation of proteoglycans. Images were analyzed using and inverted microscope.
Intravenous injection of 14C-labeled AD-MSCs in nude mice
BALB/c nude mice (6 weeks old, male, weight 19–22 g) were supplied by Orient Bio (Seongnam, Republic of Korea). All experimental procedures were performed according to the guidelines and with the approval of the Institutional Animal Care and Use Committee (IACUC) of the Korea Institute of Science and Technology (IACUC number: KIST-2019–002). Housed at the Integrated Animal Center of the Korea Institute of Science and Technology (KIST) maintained on a 12 h interval day and night cycle. Mice were allowed free access to water and feed. In order to confirm the distribution of 14C-labeled AD-MSCs after the transplantation of cells into mice, five different groups of mice were prepared for assessment at 4 h, 12 h, 24 h, 48 h and 7 days after intravenous (IV) injection (n = 3). Then, close to 1 \(\times\) 106 of 14C-labeled AD-MSCs suspended in 150 μl of PBS were injected into each mouse via the tail vein. After the injection of the 14C-labeled AD-MSCs, the mice were sacrificed with CO2 and organs (lung, spleen, liver, heart, kidney, brain) were extracted. In order to prevent 14C contamination between organs during separation, each part was separated a set of forceps that underwent frequent cleaning them with PBS. The extracted organs were washed with PBS and then frozen in a cryo-tube. Samples were stored in a -80 °C until further processing. All animal experiments were approved by an ethics committee organized in KIST.
Graphitization of samples and accelerator mass spectrometry (AMS) measurement
In order to homogenize collected organ were washed 2 times in PBS prior to organ homogenization using a BeadBug microtube homogenizer (Benchmark scientific, Edison, NJ, USA) in TEN buffer. After homogenization, about ~ 25 μl of each organ sample were dried using vacuum concentrator (LABCONCO, Kansas City, Mo, USA) and copper oxide (600 mg) were added to the sample tube followed by placing into a quartz tube for oxidation. Briefly, dried sample were sealed by the propane-oxygen flame under vacuum and oxidized at 900 °C for 3 h in muffle furnace (JISICO, Seoul, Republic of Korea). The oxidized gases (importantly isotopologues of gaseous carbon dioxide) were transferred to an in situ reduction tube via cryogenic trapping by transfer system manufactured at KIST; the non-condensable gases at liquid nitrogen temperature (-196 °C) were removed under vacuum and the carbon dioxide(s) reacted at 530 °C for 6 h to form graphite over the iron catalyst26. The graphite generated on the iron powder was mixed using a metal rod and then pressed using a KIST pressing tool to generate the target for AMS measurement. The KIST AMS is a 6MV Tandentron (HVEE, High Voltage Engineering Europa, BV, The Netherlands) with a terminal voltage set to 3 MV to accelerate carbon atomic ions(C-);the system background was 14C/12C = ~ 5 \(\times\) 10–16 (as determined using pyrolytic graphite sheet). SRM 4990C (1.3407 Modern Carbon; Oxalic acid II) standard sample provided by National Institute of Standards and Technology (NIST) were prepared and measured for the calibration of AMS samples. C3 (1.2941 Modern carbon, cellulose) and C8 (0.1503 Modern carbon, oxalic acid) as reference materials (obtained from IAEA) were also prepared for the confirming sample treatment and AMS measurement process. In general at least 50,000 “counts” were acquired to obtain under under-1% imprecision. The measured 14C/12C ratios of each sample were normalized to the Oxalic acid II results and the final Modern carbon values were calculated.
Detection of DiI labeled AD-MSCs in nude mice
The organs were dehydrate in 4% PFA with a graded sucrose series then embedded in OCT compound to obtain 5 µm cryo section. This was used to identify AD-MSCs with DiI staining.
Statistical analysis
Statistical analyses were performed using the by GraphPad Prism software (La Jolla, CA, USA; Version 5). Data are presented as mean ± SD, and the statistical significance of the experimental results were calculated using one-way ANOVA. A value of p < 0.05 was considered statistically significant.
References
Chen, G. et al. In vivo real-time visualization of mesenchymal stem cells tropism for cutaneous regeneration using NIR-II fluorescence imaging. Biomaterials 53, 265–273. https://doi.org/10.1016/j.biomaterials.2015.02.090 (2015).
MacAskill, M. G. et al. Robust revascularization in models of limb ischemia using a clinically translatable human stem cell-derived endothelial cell product. Mol. Ther. J. Am. Soc. Gene Ther. 26, 1669–1684. https://doi.org/10.1016/j.ymthe.2018.03.017 (2018).
Nam, S. Y., Chung, E., Suggs, L. J. & Emelianov, S. Y. Combined ultrasound and photoacoustic imaging to noninvasively assess burn injury and selectively monitor a regenerative tissue-engineered construct. Tissue Eng. Part C Methods 21, 557–566. https://doi.org/10.1089/ten.TEC.2014.0306 (2015).
Ricles, L. M., Nam, S. Y., Trevino, E. A., Emelianov, S. Y. & Suggs, L. J. A dual gold nanoparticle system for mesenchymal stem cell tracking. J. Mater. Chem. B 2, 8220–8230. https://doi.org/10.1039/C4TB00975D (2014).
Zhang, L. et al. High MRI performance fluorescent mesoporous silica-coated magnetic nanoparticles for tracking neural progenitor cells in an ischemic mouse model. Nanoscale 5, 4506–4516. https://doi.org/10.1039/c3nr00119a (2013).
Cheng, S. H. et al. Dynamic in vivo SPECT imaging of neural stem cells functionalized with radiolabeled nanoparticles for tracking of glioblastoma. J. Nucl. Med. Off. Publ. Soc. Nuclear Med. 57, 279–284. https://doi.org/10.2967/jnumed.115.163006 (2016).
Lee, S. et al. In vivo stem cell tracking with imageable nanoparticles that bind bioorthogonal chemical receptors on the stem cell surface. Biomaterials 139, 12–29. https://doi.org/10.1016/j.biomaterials.2017.05.050 (2017).
Jeong, H. J. et al. Macrophage cell tracking PET imaging using mesoporous silica nanoparticles via in vivo bioorthogonal F-18 labeling. Biomaterials 199, 32–39. https://doi.org/10.1016/j.biomaterials.2019.01.043 (2019).
Kim, Y. S. et al. Tracking intravenous adipose-derived mesenchymal stem cells in a model of elastase-induced emphysema. Tuberc. Respir. Dis. 77, 116–123. https://doi.org/10.4046/trd.2014.77.3.116 (2014).
Prigent, J. et al. Human progenitor cell quantification after xenotransplantation in rat and mouse models by a sensitive qPCR assay. Cell Transplant. 24, 1639–1652. https://doi.org/10.3727/096368914x681955 (2015).
Vander Borght, T. M., Lambotte, L. E., Pauwels, S. A. & Dive, C. C. Uptake of thymidine labeled on carbon 2: a potential index of liver regeneration by positron emission tomography. Hepatology 12, 113–118. https://doi.org/10.1002/hep.1840120118 (1990).
Wu, C. G. et al. In vivo tracking of human placenta derived mesenchymal stem cells in nude mice via (1)(4)C-TdR labeling. BMC Biotechnol. 15, 55. https://doi.org/10.1186/s12896-015-0174-4 (2015).
Hampe, D., Gleisberg, B., Akhmadaliev, S., Rugel, G. & Merchel, S. Determination of 41Ca with LSC and AMS: method development, modifications and applications. J. Radioanal. Nuclear Chem. 296, 617–624. https://doi.org/10.1007/s10967-012-2145-8 (2013).
Lappin, G. & Garner, R. C. Big physics, small doses: the use of AMS and PET in human microdosing of development drugs. Nat. Rev. Drug Discov. 2, 233–240. https://doi.org/10.1038/nrd1037 (2003).
Xu, X. et al. Overcoming bioanalytical challenges in an Onglyza® intravenous [14C]microdose absolute bioavailability study with accelerator MS. Bioanalysis 4, 1855–1870. https://doi.org/10.4155/bio.12.171 (2012).
Hum, N. R. et al. Tracking tumor colonization in xenograft mouse models using accelerator mass spectrometry. Sci. Rep. 8, 15013. https://doi.org/10.1038/s41598-018-33368-0 (2018).
Mukherjee, A. et al. Radiolabeling of umbilical cord-derived mesenchymal stem cells for in vivo tracking. Cancer Biother. Radiopharm. 27, 614–619. https://doi.org/10.1089/cbr.2011.1146 (2012).
Detante, O. et al. Intravenous administration of 99mTc-HMPAO-labeled human mesenchymal stem cells after stroke: in vivo imaging and biodistribution. Cell Transplant. 18, 1369–1379. https://doi.org/10.3727/096368909x474230 (2009).
Feng, S. W. et al. Dynamic distribution of bone marrow-derived mesenchymal stromal cells and change of pathology after infusing into mdx mice. Cytotherapy 10, 254–264. https://doi.org/10.1080/14653240802020381 (2008).
Kelly, S. J. & Rossant, J. The effect of short-term labelling in (3H) thymidine on the viability of mouse blastomeres: alone and in combination with unlabelled blastomeres. J. Embryol. Exp. Morphol. 35, 95–106 (1976).
Keprtova, J. & Minarova, E. The effect of 3H-thymidine on the proliferation of in vitro cultured mammalian cells. Gen. Physiol. Biophys. 4, 81–92 (1985).
Gholamrezanezhad, A. et al. In vivo tracking of 111In-oxine labeled mesenchymal stem cells following infusion in patients with advanced cirrhosis. Nuclear Med. Biol. 38, 961–967. https://doi.org/10.1016/j.nucmedbio.2011.03.008 (2011).
Liu, G. et al. Tracking of transplanted human umbilical cord-derived mesenchymal stem cells labeled with fluorescent probe in a mouse model of acute lung injury. Int. J. Mol. Med. 41, 2527–2534. https://doi.org/10.3892/ijmm.2018.3491 (2018).
Caplan, H. et al. Mesenchymal stromal cell therapeutic delivery: translational challenges to clinical application. Front Immunol. 10, 1645. https://doi.org/10.3389/fimmu.2019.01645 (2019).
Jeong, Y. H. et al. The effect of hexanoyl glycol chitosan on the proliferation of human mesenchymal stem cells. Polymers https://doi.org/10.3390/polym10080839 (2018).
Kim, J. et al. Progress report of the innovated KIST ion beam facility. Nuclear Instrum. Methods Phys. Res. Sect. B Beam Interact. Mater. Atoms. https://doi.org/10.1016/j.nimb.2016.10.038 (2016).
Acknowledgements
This research was supported by intramural funding from the Korea Institute of Science and Technology (2V06990). This work was supported by the National Research Foundation of Korea (NRF) grants from the Ministry of Science, ICT (No. 2015-M3A9C7030091) and by grant from the KRIBB Research Initiative Program.
Author information
Authors and Affiliations
Contributions
B.Y.Y. and H.M.C. supervised the study, generated the figures, and wrote the manuscript. M.S.O. and S.G.L. performed experiments, generated the figures, and wrote the manuscript. G.H.L. generated data, and provided important input to the experimental design. C.Y.K. wrote the manuscript, and provided important input to the experimental design. E.Y.K. and J.H.S. generated data.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Oh, MS., Lee, SG., Lee, GH. et al. In vivo tracking of 14C thymidine labeled mesenchymal stem cells using ultra-sensitive accelerator mass spectrometry. Sci Rep 11, 1360 (2021). https://doi.org/10.1038/s41598-020-80416-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-80416-9
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.