Abstract
The correlation between G388R or V10I polymorphisms of fibroblast growth factor receptor (FGFR) 4 gene and the risk of carcinoma has been investigated previously, but the results are contradictory. Odds ratios (ORs) with 95% confidence intervals (95%CIs), in silico tools, and immunohistochemical staining (IHS) were adopted to assess the association. In total, 13,793 cancer patients and 16,179 controls were evaluated in our pooled analysis. Summarization of all the studies showed that G388R polymorphism is associated with elevated susceptibility to cancer under homozygous comparison (OR = 1.21, 95%CI = 1.03–1.43, P = 0.020) and a recessive genetic model (OR = 1.21, 95%CI = 1.04–1.41, P = 0.012). In the stratification analysis by cancer type and ethnicity, similar findings were indicated for prostate cancer, breast cancer, and individuals of Asian descendant. Polyphen2 bioinformatics analysis showed that the G388R mutation is predicted to damage the protein function of FGFR4. IHS analysis indicated that FGFR4 expression is increased in advanced prostate cancer. These findings may guide personalized treatment of certain types of cancers. Up-regulation of FGFR4 may be related to a poor prognosis in prostate cancer.
Similar content being viewed by others
Introduction
Cancer remains a global threat to public health and poses a huge economic burden on the societies of both developing and developed countries. Breast cancer, colorectal cancer (CRC), prostate cancer, hepatocellular carcinoma (HCC), and head and neck squamous cell carcinoma (HNSCC) are the most common cancers in the world. However, the etiology underlying cancer development is far from comprehensively demonstrated1. Gene mutations, such as single nucleotide polymorphism (SNP), have been indicated in recent years to have an impact on the susceptibility of cancer2,3.
Fibroblast growth factor receptor 4 (FGFR4) is a member of the family of fibroblast growth factor receptors. It displays a variety of biological activities, including angiogenic and mitogenic activities. It can also transduce signals of more than 20 known ligands, such as those involved in cell proliferation, differentiation and development4,5. FGFR4 is highly activated in many types of solid tumors and hematological malignancies in which it drives the development and progression of cancer as an oncogene6. In addition, immunohistochemical evaluations have shown that the strong expression of FGFR4 in malignant tumor cells is significantly correlated with an increase in clinical stage and tumor grade and a decrease in patients' survival rates7.
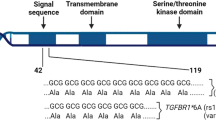
Recently, several studies have focused on the relationship between FGFR4 gene variant and susceptibility of cancer8,9,10,11,12. A common SNP, rs351855, which leads to substitution of glycine by arginine at codon 388 in the domain of the FGFR4 receptor (Gly388Arg, G388R), has been reported to be associated with cancer risk. In 2017, a meta-analysis assessed FGFR4 G388R polymorphism and found that it was correlated with an elevated susceptibility of several cancers8. Since then, more case–control studies have been conducted. Nevertheless, the correlation between FGFR4 G388R, V10I variants, and susceptibility to carcinoma remains controversial. To comprehensively investigate the relationship between FGFR4 G388R, V10I variants and cancer risk, we conducted the present analysis based on all eligible studies and used online databases and immunohistochemical staining (IHS) to assess the expression of FGFR4 further9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33.
Methods
Literature identification
A comprehensive search of eligible publications related to FGFR4 G388R or V10I polymorphisms and cancer risk was performed using PubMed, EMbase, and the Chinese Wanfang database. The search terms were: (‘FGFR4’ OR ‘fibroblast growth factor receptor 4’) AND (‘variant’ OR ‘mutation’ OR ‘polymorphism’) AND (‘cancer’ OR ‘carcinoma’ OR ‘malignant tumor’). The latest search was conducted on May 2, 2020. The references of related reviews and original articles, as well as supplementary material, were also evaluated to maximize the coverage of the present analysis.
Inclusion criteria and exclusion criteria
The inclusion criteria were as follows: (a) investigations of the relationship between FGFR4 G388R or V10I polymorphisms and risk of cancer; (b) cohort or case–control studies; (c) sufficient genotype information to calculate Odds ratios (ORs) and 95% confidence intervals (95%CIs); and (d) P-values greater than 0.05 for Hardy–Weinberg equilibrium (HWE) of controls. Articles that departed from HWE were removed. We also excluded studies with no control population. When repeated studies appeared, only the latest or largest articles were included.
Data extraction
Two authors independently searched the articles and extracted data from individual studies according to the inclusion criteria. Information collected from all eligible studies included the name of the first author, publication date, the ethnicity of subjects in the study, source of control, number of genotyped cases and controls, P-values for HWE of controls, and genotyping method. If a type of cancer appeared in only one study, then this cancer was classified in to ‘other cancer’ group. A total of 37 eligible studies were included.
Statistical analysis
We adopted ORs with 95% CI to explore the correlation between FGFR4 G388R or V10I polymorphisms and the risk of cancer. For the G388R variant, five genetic models were used (allelic contrast, R vs. G, heterozygous model, RG vs. GG, homozygous model, RR vs. GG; dominant model, RR + RG vs. GG, recessive model RR vs. RG + GG). For V10I polymorphism, the five models were as follows: I versus V; IV versus VV; II versus VV; II + IV versus VV; II versus IV + VV. The homogeneity of the study was calculated by a chi-square-based Q-test. P-value ≥ 0.05 indicated a lack of heterogeneity; the summary OR was evaluated by the fixed-effects model (Mantel–Haenszel method). Otherwise, the random-effects model was employed. Begg's funnel plot and sensitivity analysis were performed to assess publication bias. Stratification analysis was applied to evaluate the impact of ethnicity and cancer type. All statistical analyses were performed using Stata software (Stata Corporation 2009. Stata Statistical Software: Release 11. College Station, TX: StataCorp LP.).
In silico and IHS analysis of FGFR4 expression
We employed an online database to assess the expression of FGFR4 in prostate cancer and normal tissues (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga). Moreover, gene–gene interaction of FGFR4 was also evaluated by an online database (http://ualcan.path.uab.edu/analysis.html). The Cancer Genome Atlas (TCGA) samples were also applied to investigate the effect of FGFR4 expression on overall survival (OS) time (http://genomics.jefferson.edu/proggene/results.php). The relationship between G388R or V10I polymorphisms and FGFR4 protein damage was analyzed by Polyphen2 tools (http://genetics.bwh.harvard.edu/pph2/).
Furthermore, we used IHS to test the tissue expression of FGFR4 in prostate cancer subjects recruited by our centers. From February 2013 to July 2018, a total of 220 patients diagnosed with prostate cancer by puncture biopsy were enrolled in our study. These patients underwent radical prostatectomy in our hospitals. Before the IHS analysis, each participant signed an informed consent. In addition to the routine pathological examination, the remaining part of the tissue removed during the operation was used for immunohistochemical examination. Paraffin-embedded samples were stained with hematoxylin and eosin to confirm cancer. Tissue sections were dewaxed in xylene, dehydrated in alcohol and washed in phosphate buffer (PBS). Each slice was incubated overnight with rabbit anti-FGFR4 monoclonal antibody at 4 °C. After being rinsed with PBS for three times, the slices were incubated with secondary antibody at 20 °C for 30 min. PBS was used instead of a primary antibody as a negative control. Two authors evaluated the prostate cancer sections separately. We investigated the intensities of FGFR4 reactivities in different samples utilizing the image J software (Version 1.45, a java-based image analysis program designed by National Institutes of Health, Bethesda, Maryland, USA, Available from: URL: https://rsb.info.nih.gov/ij/) (range from score 1 to 8)34,35. The present study was approved by Ethics Committee of Changzhou No.2 People's Hospital and Affiliated Hospital of Jiangnan University.
Ethical approval and consent to participate
The present research was approved by Ethics Committee of Changzhou No.2 People's Hospital and Affiliated Hospital of Jiangnan University.
Results
Characteristics of included studies
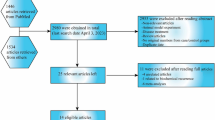
A total of 269 articles were initially involved based on the inclusion criteria (Supplement Fig. 1). After screening the abstracts, we excluded 95 articles. Then, another 149 articles were removed because they were reviews, duplicates, or studies with no control group or focus on other SNPs. Finally, 25 eligible articles (with 37 studies) concerning FGFR4 G388R or V10I polymorphisms were included in our pooled analysis. Data were collected on 29,972 participants (13,793 cancer subjects and 16,179 controls) from 30 case–control studies on FGFR4 G388R polymorphism. The most common types of cancer were prostate cancer (6 studies, n = 4610), breast cancer (6 studies, n = 3008), hepatocellular carcinoma (HCC) (3 studies, n = 2481), oral squamous cell carcinoma (OSCC) (2 studies, n = 2396), colorectal cancer (CRC) (3 studies, n = 2349), lung cancer (2 studies, n = 899), cervical cancer (2 studies, n = 885) and other cancers (6 studies, n = 3975) (Table 1). Subgroup analysis by ethnicity evaluated 15 studies on European populations, 13 studies on Asians, one study on Latin Americans, and one study on Africans. For the FGFR4 V10I variant, seven studies with 9369 subjects (4377 cases and 4992 controls) in total were identified, and of these, four studies focused on Asians (n = 5055), two on Europeans (n = 4087), and one on Africans (n = 227). In the stratification analysis by cancer type, two studies concentrated on prostate cancer. There was only one study each on cervical cancer, OSCC, breast cancer, HCC, and skin cancer. Furthermore, we checked the minor allele frequencies (MAF) of the worldwide population reported from gnomAD database. For FGFR4 G388R polymorphism, the frequency in the global population was 0.327; African descendants, 0.131; Americans, 0.420; Asians, 0.389; Europeans, 0.294; Ashkenazi Jewish, 0.320; and others, 0.318 (Fig. 1A). For the FGFR4 V10I variant, the frequency in the global population was 0.246; Africans, 0.051; Americans, 0.319; Asians, 0.332; Europeans, 0.228; Ashkenazi Jewish, 0.159; and others, 0.213 (Fig. 1B).
Main results
The overall results showed that the FGFR4 G388R variant was associated with elevated susceptibility to cancer under homozygous comparison (OR = 1.21, 95%CI = 1.03–1.43, Pheterogeneity < 0.001, P = 0.020) and recessive genetic modeling (OR = 1.21, 95%CI = 1.04–1.41, P value for heterogeneity < 0.001, P = 0.012, Table 2). The stratification analysis by cancer type revealed that individuals with the RR + RG allele had a 1.20-fold higher susceptibility to prostate cancer than those with the GG allele (95%CI = 1.06–1. 35, Pheterogeneity = 0.892, P = 0.004, Fig. 2A). Individuals with the RR + RG allele had a 1.26-fold higher risk of breast cancer than those with the wild type (95%CI = 1.14–1.54, Pheterogeneity = 0.197, P < 0.001). In subgroup analysis by ethnicity, we observed that Asian descendants carrying the RR allele had a 1.28-fold increased risk of cancer compared with those carrying the RG + GG allele (95%CI = 1.02–1.60, Pheterogeneity < 0.001, P = 0.034, Fig. 3A). However, we did not identify positive results in European (95%CI = 0.93–1.26, P = 0.306), African (95%CI = 0.13–5.00, P = 0.828), or Latin Americans (95%CI = 0.25–6.46, P = 0.780). For FGFR4 V10I polymorphism, no significant correlation was found when all studies were pooled (I vs. V, OR = 0.94, 95%CI = 0.85–1.04, P value for heterogeneity = 0.049, P = 0.227; IV vs. VV, OR = 0.97, 95%CI = 0.89–1.07, Pheterogeneity = 0.169, P = 0.601; II vs. VV, OR = 0.92, 95%CI = 0.72–1.17, P value for heterogeneity = 0.020, P = 0.488; II + IV vs. VV, OR = 0.95, 95%CI = 0.87–1.04, P value for heterogeneity = 0.147, P = 0.300, Fig. 2B; II vs. IV + VV, OR = 0.90, 95%CI = 0.74–1.11, P value for heterogeneity = 0.020, P = 0.328). Similar findings were indicated in the subgroup analysis by cancer type. In stratification analysis by ethnicity, we observed that FGFR4 V10I mutation may not have an impact on the risk of cancer for Asian (95%CI = 0.66–1.08, P = 0.184, Fig. 3B), African (95%CI = 0.34–1.86, P = 0.598), or individuals with European descent (95%CI = 0.83–1.42, P = 0.563).
In silico and IHS analyses of FGFR4 expression
We used in silico tools to investigate whether G388R and V10I mutations affect the protein function of FGFR4. Polyphen2 bioinformatics analysis showed that FGFR4 G388R was predicted to damage protein function, with a score of 0.700 (Fig. 4A). However, the V10I variation was predicted to be benign, with a score less than 0.001 (Fig. 4B). We also utilized an online database to assess the expression of FGFR4 in prostate cancer participants and normal controls. As described in Fig. 5A, FGFR4 expression is elevated in prostate cancer compared with that in the control. TCGA samples were also analyzed to investigate the effect of FGFR4 expression on OS time. No significant difference in OS time was observed between the high FGFR4 expression group and the low expression group (P > 0.05, Fig. 5B).
In order to demonstrate the expression of FGFR4 in prostate cancer tissues, we applied IHS to evaluate its expression among prostate cancer patients at our centers. A total of 220 prostate cancer participants were enrolled in our centers. The feature distribution from prostate cancer volunteers has been provided in our previous article36. Immunohistochemistry of FGFR4 in prostate cancer specimens is described in Fig. 6. The intensity of immunoreactivity was mainly concentrated in the cytoplasm of prostate cancer epithelial cells (Fig. 6C,D). The expression of FGFR4 was up-regulated in more advanced cases (Fig. 6D) compared with early stage cases (Fig. 6A,B, P < 0.05). Moreover, the gene–gene correlation of FGFR4 was also assessed. At least 24 genes were shown to participate in interactions with FGFR4 (Fig. 7A). The most related genes to FGFR4 include: CORIN (corin, serine peptidase, Fig. 7B), NKD1 (Naked1, NKD inhibitor 1, Fig. 7C), and CALML3 (calmodulin like 3, Fig. 7D).
Tissue expression of FGFR4 among prostate cancer participants. The intensity of immunoreactivity was mainly concentrated in the cytoplasm of prostate cancer epithelial cells (C,D). The expression of FGFR4 is up-regulated in more advanced cases (D), as compared to ones in early stage (A,B, P < 0.05).
Gene–gene interaction of FGFR4 in prostate cancer (A). Correlation analysis by TCGA samples show that CORIN (corin, serine peptidase, B) is predicted to positively correlated with FGFR4. The interaction between NKD1 (Naked1, NKD inhibitor of WNT signaling pathway 1) and FGFR4 is shown in (C). The correlation between CALML3 (calmodulin like 3) and FGFR4 is described in (D).
Publication bias and sensitivity analysis
A Begg's funnel plot was employed to investigate publication bias. No evidence of asymmetry was observed for FGFR4 G388R (t = − 1.52, P = 0.140, Fig. 8A) or V10I variants (t = 0.07, P = 0.945, Fig. 8B). Sensitivity analysis of FGFR4 G388R or V10I polymorphisms and the risk of cancer was performed by removing individual studies in turn. No single study influenced the overall OR, indicating that the results of the above analysis for FGFR4 G388R (Fig. 8C) and V10I (Fig. 8D) polymorphisms are reliable.
Discussion
The etiology of cancer has not been totally elucidated. Clinically, SNPs have various influences on the development of diseases, including cancer37,38,39. The association between FGFR4 G388R or V10I variants and the susceptibility of cancer has been evaluated previously, but the results are conflicting. For example, Ma et al. performed a case–control study and found that the R-allele of the G388R variant had a significant impact on the development and progression of prostate cancer in Japanese patients27. However, this conclusion could not be confirmed by the research of FitzGerald et al., who observed no positive correlation between FGFR4 G388R or V10I polymorphisms and prostate cancer susceptibility in Caucasians or African Americans23. Xiong et al. performed a meta-analysis using articles published before October 2016 to assess the effect of the FGFR4 G388R variant8. They showed that the G388R variant was correlated with increased susceptibility to prostate and breast cancer and reduced risk of lung cancer. In our analysis, all eligible studies based on inclusion criteria were included to extensively evaluate the association between FGFR4 G388R or V10I variants and the susceptibility of cancer. We further adopted in silico and IHS analysis to confirm the above conclusion.
We performed a pooled analysis of studies that included 9416 cancer participants and 11,187 control subjects to investigate the relationship between the FGFR4 G388R variant and susceptibility to cancer. In the current analysis, we found that G388R polymorphism is associated with an elevated risk of cancer. Furthermore, stratifying by type of cancer, we observed that this variant is correlated with prostate and breast cancer, but not with lung cancer. Our results are consistent with those of Wei et al. and Xu et al.37,40. In subgroup analysis by ethnicity, we also found that the FGFR4 R-allele is correlated with an increased risk of cancer in individuals with Asian descent. For FGFR4 V10I polymorphism, no significant relationship was indicated in either overall or stratifying analysis. The conclusions derived from our analysis were consistent with a previous meta-analysis published in 201041. In 2017, another meta-analysis found that the FGFR4 388R variation was a reduced risk factor for lung cancer8. However, we did not come to this conclusion in the current analysis. The reason may be that there were few studies in our analysis that were focused on lung cancer15,33. Therefore, further well-designed studies with large sample sizes are needed to confirm the role of FGFR4 G388R polymorphisms in lung cancer in future. Furthermore, we employed an in silico tool to investigate whether the G388R and V10I mutations could affect the protein function of FGFR4. It showed that the G388R mutation, but not the V10I mutation, could damage the protein function of FGFR4. We further utilized TCGA samples to assess the expression of FGFR4 in prostate cancer participants. The FGFR4 expression was elevated in prostate cancer compared with that in the control group. Nevertheless, no significant difference in the OS time could be identified between the high FGFR4 expression group and the low expression group. In addition, we applied IHS to evaluate its expression among prostate cancer subjects in our centers. The expression of FGFR4 is increased in more advanced cases, which indicates that up-regulation of FGFR4 is related to a poor prognosis of prostate cancer.
There are some potential limitations in the present analysis. First, the P-value of heterogeneity was less than 0.05 in five genetic models when all studies were pooled to assess FGFR4 G388R polymorphism. Although the Der Simonian and Laird method (random-effect model) was used42, the analysis may have been influenced by potential bias. Second, the number of eligible studies on the FGFR4 V10I variant in the present analysis remains insufficient for a comprehensive analysis. In our subgroup analysis by cancer type, only two studies concentrated on prostate cancer. A very limited number of studies were available for multiple types of cancers such as cervical cancer, OSCC, breast cancer, HCC, and skin cancer. Further research including more participants with various carcinomas is warranted to confirm this effect. Third, a previous study demonstrated that FGFR4 G388R polymorphism was related to up-regulation of FGFR4 in breast cancer41. For prostate cancer, further in vitro experiments are required to confirm the results from our pooled analysis. More functional research is warranted to determine whether the G388R mutation is responsible for increased expression of FGFR4. Moreover, genotyping the same patients will provide more persuasive evidence of a correlation between genotype or alleles and tissue expression of FGFR4 observed by IHS analysis. Finally, at least 24 genes are involved in the interaction with FGFR4. Since few studies on these specific relationships can be retrieved from an online database, future studies are needed to ascertain these correlations in more detail.
In summary, our study showed that FGFR4 G388R polymorphism is associated with an elevated risk of cancer, especially for prostate and breast cancer. The R-allele of the FGFR4 G388R variant is correlated with an increased risk of cancer in individuals with Asian descent. The G388R mutation, but not the V10I mutation, is predicted to damage the protein function of FGFR4. Up-regulation of FGFR4 may be related to a poor prognosis in prostate cancer. These findings may guide personalized treatment of certain types of cancers.
Data availability
All the data generated in the above research can be acquired from the corresponding authors upon reasonable request. All methods were conducted in accordance with relevant guidelines and regulations.
Abbreviations
- HNSCC:
-
Head and neck squamous cell carcinoma
- HCC:
-
Hepatocellular carcinoma
- OSCC:
-
Oral squamous cell carcinoma
- CRC:
-
Colorectal cancer
- HWE:
-
Hardy–Weinberg equilibrium of controls
- HB:
-
Hospital-based
- LA:
-
Latin Americans
- NA:
-
Not available
- NHL:
-
Non-Hodgkin's lymphoma
- PB:
-
Population-based
- PCR–RFLP:
-
Polymerase chain reaction and restrictive fragment length polymorphism
References
Torre, L. A. et al. Global cancer statistics, 2012. CA Cancer J. Clin. 65, 87–108 (2015).
Shao, H. B. et al. Human methionine synthase A2756G polymorphism increases susceptibility to prostate cancer. Aging (Albany NY) 10, 1776–1788 (2018).
Zhang, L. F. et al. VEGF gene rs3025039C/T and rs833052C/A variants are associated with bladder cancer risk in Asian descendants. J. Cell Biochem. 120, 10402–10412 (2019).
Wilkie, A. O., Morriss-Kay, G. M., Jones, E. Y. & Heath, J. K. Functions of fibroblast growth factors and their receptors. Curr. Biol. 5, 500–507 (1995).
Powers, C. J., McLeskey, S. W. & Wellstein, A. Fibroblast growth factors, their receptors and signaling. Endocr. Relat. Cancer 7, 165–197 (2000).
Tang, S., Hao, Y., Yuan, Y., Liu, R. & Chen, Q. Role of fibroblast growth factor receptor 4 in cancer. Cancer Sci. 109, 3024–3031 (2018).
Gowardhan, B. et al. Evaluation of the fibroblast growth factor system as a potential target for therapy in human prostate cancer. Br. J. Cancer 92, 320–327 (2005).
Xiong, S. W. et al. Functional FGFR4 Gly388Arg polymorphism contributes to cancer susceptibility: evidence from meta-analysis. Oncotarget 8, 25300–25309 (2017).
Wimmer, E. et al. Fibroblast growth factor receptor 4 single nucleotide polymorphism Gly388Arg in head and neck carcinomas. World J. Clin. Oncol. 10, 136–148 (2019).
Chen, T. H. et al. Association of fibroblast growth factor receptor 4 genetic polymorphisms with the development of uterine cervical cancer and patient prognosis. Reprod. Sci. 25, 86–93 (2018).
Li, Y. P., Zhang, L., Zou, Y. L. & Yu, Y. Association between FGFR4 gene polymorphism and high-risk HPV infection cervical cancer. Asian Pac. J. Trop. Med. 10, 680–684 (2017).
Chou, C. H. et al. Functional FGFR4 Gly388Arg polymorphism contributes to oral squamous cell carcinoma susceptibility. Oncotarget 8, 96225–96238 (2017).
Sheu, M. J. et al. Fibroblast growth factor receptor 4 polymorphism is associated with liver cirrhosis in hepatocarcinoma. PLoS ONE 10, e0122961 (2015).
Jiang, Y. et al. Association of FGFR3 and FGFR4 gene polymorphisms with breast cancer in Chinese women of Heilongjiang province. Oncotarget 6, 34023–34029 (2015).
Ture, M. et al. Investigation of FGFR4 (Gly388Arg) gene polymorphism in primary lung cancer patients. Int. J. Hum. Genet. 15, 7–12 (2015).
Gao, L. et al. Fibroblast growth factor receptor 4 polymorphism is associated with increased risk and poor prognosis of non-Hodgkin’s lymphoma. Tumour. Biol. 35, 2997–3002 (2014).
Shen, Y. Y. et al. Fibroblast growth factor receptor 4 Gly388Arg polymorphism in Chinese gastric cancer patients. World J. Gastroenterol. 19, 4568–4575 (2013).
Heinzle, C. et al. Differential effects of polymorphic alleles of FGF receptor 4 on colon cancer growth and metastasis. Cancer Res. 72, 5767–5777 (2012).
Yang, Y. et al. Association between fibroblast growth factor receptor 4 polymorphisms and risk of hepatocellular carcinoma. Mol. Carcinog. 51, 515–521 (2011).
Batschauer, A. P. et al. HFE, MTHFR, and FGFR4 genes polymorphisms and breast cancer in Brazilian women. Mol. Cell Biochem. 357, 247–253 (2011).
Ho, C. K., Anwar, S., Nanda, J. & Habib, F. K. FGFR4 Gly388Arg polymorphism and prostate cancer risk in Scottish men. Prostate Cancer Prostatic Dis. 13, 94–96 (2010).
Tanuma, J. et al. FGFR4 polymorphism, TP53 mutation, and their combinations are prognostic factors for oral squamous cell carcinoma. Oncol. Rep. 23, 739–744 (2010).
FitzGerald, L. M. et al. Association of FGFR4 genetic polymorphisms with prostate cancer risk and prognosis. Prostate Cancer Prostatic Dis. 12, 192–197 (2009).
Ho, H. K. et al. Fibroblast growth factor receptor 4 regulates proliferation, anti-apoptosis and alpha-fetoprotein secretion during hepatocellular carcinoma progression and represents a potential target for therapeutic intervention. J. Hepatol. 50, 118–127 (2009).
Naidu, R., Har, Y. C. & Taib, N. A. Polymorphism of FGFR4 Gly388Arg does not confer an increased risk to breast cancer development. Oncol. Res. 18, 65–71 (2009).
Nan, H., Qureshi, A. A., Hunter, D. J. & Han, J. Genetic variants in FGFR2 and FGFR4 genes and skin cancer risk in the Nurses’ Health Study. BMC Cancer 9, 172 (2009).
Ma, Z. et al. Polymorphisms of fibroblast growth factor receptor 4 have association with the development of prostate cancer and benign prostatic hyperplasia and the progression of prostate cancer in a Japanese population. Int. J. Cancer 123, 2574–2579 (2008).
Mawrin, C. et al. Analysis of a single nucleotide polymorphism in codon 388 of the FGFR4 gene in malignant gliomas. Cancer Lett. 239, 239–245 (2006).
Spinola, M. et al. FGFR4 Gly388Arg polymorphism and prognosis of breast and colorectal cancer. Oncol. Rep. 14, 415–419 (2005).
Wang, J., Stockton, D. W. & Ittmann, M. The fibroblast growth factor receptor-4 Arg388 allele is associated with prostate cancer initiation and progression. Clin. Cancer Res. 10, 6169–6178 (2004).
Morimoto, Y. et al. Single nucleotide polymorphism in fibroblast growth factor receptor 4 at codon 388 is associated with prognosis in high-grade soft tissue sarcoma. Cancer 98, 2245–2250 (2003).
Bange, J. et al. Cancer progression and tumor cell motility are associated with the FGFR4 Arg(388) allele. Cancer Res. 62, 840–847 (2002).
Spinola, M. et al. Functional FGFR4 Gly388Arg polymorphism predicts prognosis in lung adenocarcinoma patients. J. Clin. Oncol. 23, 7307–7311 (2005).
Wirawati, V. et al. The distribution of serotonergic nerve on the hippocampus of the fruit bats (Rousettus amplexicaudatus). Vet. World 12, 1460–1466 (2019).
Stanchev, S. et al. Differential collagen expression in kidney and heart during hypertension. Bratisl. Lek. Listy. 121, 73–78 (2020).
Zhang, L. F. et al. Association between SOD2 V16A variant and urological cancer risk. Aging (Albany NY) 12, 825–843 (2020).
Wei, W. et al. Prognostic implications of fibroblast growth factor receptor 4 polymorphisms in primary breast cancer. Mol. Carcinog. 7, 988–996 (2018).
Dai, F. et al. The association between three AXIN2 variants and cancer risk. J. Cell Biochem. 120, 15561–15571 (2019).
Zhu, L. et al. CDKN1B Val 109 Gly variant is not related to risk of prostate cancer. J. Cell Biochem. 120, 18346–18356 (2019).
Xu, B. et al. FGFR4 Gly388Arg polymorphism contributes to prostate cancer development and progression: a meta-analysis of 2618 cases and 2305 controls. BMC Cancer 11, 84 (2011).
Xu, W. et al. FGFR4 transmembrane domain polymorphism and cancer risk: a meta-analysis including 8555 subjects. Eur. J. Cancer 46, 3332–3338 (2010).
DerSimonian, R. & Laird, N. Meta-analysis in clinical trials. Control Clin. Trials 7, 177–188 (1986).
Funding
This study was supported by funding from Young Talent Development Plan of Changzhou Health Commission (No. CZQM2020065), Changzhou No.2 People's Hospital Young Scientists Foundation (2019K008).
Author information
Authors and Affiliations
Contributions
Y.M., L.Z., and L.F.Z. designed the study; Methodology, Z.Z., and X.Z.; W.Z., T.P., and W.Y. collected the data; Formal analysis, H.W., Q.S., and L.S.; Z.L., Y.M. and L.F.Z. revised the manuscript; Z.Z. and L.F.Z. wrote the manuscript. All the authors approved the final edition of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Peng, T., Sun, Y., Lv, Z. et al. Effects of FGFR4 G388R, V10I polymorphisms on the likelihood of cancer. Sci Rep 11, 1373 (2021). https://doi.org/10.1038/s41598-020-80146-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-80146-y
This article is cited by
-
Predictive significance of FGFR4 p.G388R polymorphism in metastatic colorectal cancer patients receiving trifluridine/tipiracil (TAS-102) treatment
Journal of Translational Medicine (2024)
-
Effect of esophageal muscle fibrosis on prognosis of per-oral endoscopic myotomy (POEM) in achalasia patients
Surgical Endoscopy (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.