Abstract
The DNA demethylating agent, 5-Azacytidine (5Aza), and histone deacetylase inhibitor, valproic acid (Vpa), can improve the reprogramming efficiencies of pluripotent cells. This study aimed to examine the roles of 5Aza and Vpa in the dedifferentiation of epithelial cell rests of Malassez (ERM) into stem-like cells. Additionally, the ability of stem-like cells to differentiate into mesenchymal cells was evaluated. ERM was cultured in embryonic stem cell medium (ESCM) with 1 µM of 5Aza, or 2 mM of Vpa, or a combination of 5Aza and Vpa. The cells stimulated with both 5Aza and Vpa were named as progenitor-dedifferentiated into stem-like cells (Pro-DSLCs). The Pro-DSLCs cultured in ESCM alone for another week were named as DSLCs. The stem cell markers were significantly higher in the DSLCs than the controls (no additions). The mRNA and protein levels of the endothelial, mesenchymal stem, and osteogenic cell markers were significantly higher in the Pro-DSLCs and DSLCs than the controls. The combination of a demethylating agent and a deacetylated inhibitor induced the dedifferentiation of ERM into DSLCs. The Pro-DSLCs derived from ERM can be directly reprogrammed into mesenchymal-like cells without dedifferentiation into stem-like cells. Isolated ERM treated with epigenetic agents may be used for periodontal regeneration.
Similar content being viewed by others
Introduction
Stem cells/progenitor cells have been isolated for use in regenerative dentistry. Several methods have been employed to purify the cells from human exfoliated deciduous teeth1. However, it is difficult to obtain cells of adequate quality and numbers; thus, a stable supply must be generated for their application in regenerative dentistry. Since the development of induced pluripotent stem (iPS) cells, several cells derived from dental cells and tissues such as the apical papillae, dental pulp, primary teeth, third molars, oral mucosa, and gingiva have been reprogrammed by transduction with Yamanaka factors, which include the Octamer-binding transcription factor 3/4 (Oct3/4), SRY-box transcription factor 2 (Sox2), Kruppel like factor 4 (Klf4), and c-Myc2,3,4. Epigenetics is the study of changes in gene expression that do not involve alterations in the gene sequences. Epigenetic alterations have been observed during the development of iPS cells5. Furthermore, the generation of reprogramming cells by inducing epigenetic alterations has been attempted recently6,7,8,9,10. DNA methylation and deacetylation are two major mechanisms of epigenetic modifications. The DNA demethylating agent (DNA methyltransferase inhibitor; DNMTi), 5-Azacytidine (5Aza), may favor reprogramming of fibroblasts, which is insufficient in the transition to pluripotency6. Moreover, 5Aza and valproic acid (Vpa), a histone deacetylase inhibitor (HDACi), have been reported to improve the reprogramming efficiencies of pluripotent stem cells7. Recently, low concentrations of HDAC were used to promote the dedifferentiation of dental pulpal stem cells11. However, epigenetic agents have not been used on other cells derived from dental tissues.
The expression of Oct3/4, Sox2, and Myc has been observed in the cells of the dental papilla and follicle12. Sox2 was localized in the dental lamina but not in the epithelial cell rests of Malassez (ERM)13. Cultured epithelial cells derived from the ERM possess stem cell properties and are capable of epithelial–mesenchymal transitions14,15. Therefore, we hypothesized that epigenetic modifications might induce the dedifferentiation of ERM with a potential for reprogramming the cells. The ERM is thought to contribute to periodontal regeneration by repairing the cementum and forming the mineralized tissue on the root surface16; thus, we presumed that the dedifferentiated ERM could be used for periodontal regeneration. In the present study, we examined the roles of the DNMTi and HDACi in dedifferentiating ERM into stem cell-like cells. In addition, the ability of these stem cell-like cells to differentiate into mesenchymal-like cells, such as endothelial, mesenchymal stem, and osteogenic cells, that constitute the periodontal ligament was evaluated.
Results
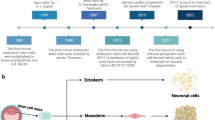
The ERM was cultured in Embryonic Stem Cell Medium (ESCM) using culture dishes under the following conditions: 1 µM of 5Aza; and/or 2 mM of Vpa; and no additions (control) for 1 week. The cells stimulated with 5Aza alone and Vpa alone were named as 5Aza1w and Vpa1w, respectively. The cells stimulated with both 5Aza and Vpa were named as a progenitor-dedifferentiated into stem-like cells (Pro-DSLCs). Some of the cells were cultured in ESCM alone for another week. The 5Aza1w and Vpa1w cultured in ESCM alone for another week were named as 5Aza2w and Vpa2w, respectively. The Pro-DSLCs cultured in ESCM alone for another week were named as dedifferentiated into stem-like cells (DSLCs). The controls (ERM cells), Pro-DSLCs, and DSLCs were further cultured in Endothelial cells (ECs) differentiation medium for 1 week, bone marrow mesenchymal stem cells (MSCs) differentiation medium for 1 week, and osteogenic differentiation medium for 3 weeks (Fig. 1).
Schematic diagram of the whole experiment. The ERM was cultured in ESCM using culture dishes under the following conditions: 1 µM of 5Aza; and/or 2 mM of Vpa; and no additions (control) for 1 week. The cells stimulated with 5Aza alone and Vpa alone were named as 5Aza1w and Vpa1w, respectively. The cells stimulated with both 5Aza and Vpa were named as a progenitor-dedifferentiated into stem-like cells (Pro-DSLCs). Some of the cells were cultured in ESCM alone for another week. The 5Aza1w and Vpa1w cultured in ESCM alone for another week were named as 5Aza2w and Vpa2w, respectively. The Pro-DSLCs cultured in ESCM alone for another week were named as a dedifferentiated into stem-like cells (DSLCs). The controls (ERM cells), Pro-DSLCs, and DSLCs were cultured in Endothelial cells (ECs) differentiation medium for 1 week, bone marrow mesenchymal stem cells (MSCs) differentiation medium for 1 week, and osteogenic differentiation medium for 3 weeks.
The cytotoxicities of different concentrations of 5Aza and Vpa were evaluated to determine the optimal concentrations. The numbers of viable ERM were significantly decreased in the groups stimulated with 10 µM of 5Aza when compared to those in the controls (no additions) at week 1, and those in the controls and 1 µM of 5Aza at week 2 (p < 0.01, 0.05; Fig. 2). The numbers of viable ERM were significantly decreased in the groups stimulated with 20 mM of Vpa at weeks 1 and 2 when compared to those in the controls (no additions) and 0.2 mM of Vpa. No significant differences in cell numbers were observed between any of the other conditions and the controls. Consequently, we decided to use 1 µM and 2 mM of 5Aza and Vpa, respectively, for further experiments.
Cell viability. The ERM was cultured at a cell density of 0.8 × 106 cells/ml (initial cells) in ESCM using 60 mm dishes under the following conditions: 0.1, 1 or 10 µM of 5Aza; and 0.2, 2 or 20 mM of Vpa; and no additions (control) for 1 week. Some of the cells were cultured in ESCM alone for another week. The numbers of viable ERM were significantly decreased in the groups stimulated with 10 µM of 5Aza when compared to those in the controls at week 1, and those in the controls and 1 µM of 5Aza at week 2 (**p < 0.01, *p < 0.05; Kruskal–Wallis test, n = 5). The numbers of viable ERM were significantly decreased in the groups stimulated with 20 mM of Vpa at weeks 1 and 2 when compared to those in the controls and 0.2 mM of Vpa. No significant differences in cell numbers were observed between any of the other conditions and the controls.
We determined the mRNA expression levels of the stem cell markers Nanog homeobox (Nanog), Oct3/4, Sox2, and Klf4, odontogenic markers amelogenin (Amel), ameloblastin (Ambn), and keratin19 (Krt19), and tumorigenic marker proliferating cell nuclear antigen (Pcna). iPS cells were used as positive controls. The expression levels of all the stem cell markers, Nanog, Oct3/4, Sox2, and Klf4, following stimulation with 1 µM of 5Aza and/or 2 mM of Vpa, were significantly higher in the dedifferentiated into stem-like cells (DSLCs) and iPS cells when compared to those in the controls (ERM cells) (p < 0.01, 0.05; Fig. 3a–d). No significant increases in the expression levels of the odontogenic markers, except for ameloblastin (Ambn), were observed in the DSLCs (Fig. 3e–g). The expression levels of Nanog were significantly upregulated in the cells treated with 5Aza alone for 1 week and followed by ESCM alone for another week (5Aza2w; p < 0.05; Fig. 3a–g). No significant increase in the expression level of any of the markers was noted in the progenitor-DSLCs (Pro-DSLCs), the cells treated with 5Aza alone for 1 week (5Aza1w), the cells treated with Vpa alone for 1 week (Vpa1w), and the cells treated with Vpa alone for 1 week and followed by ESCM alone for another week (Vpa2w; Fig. 3a–g). No significant increase in the expression level of Pcna was observed under any of the culture conditions (Fig. 3h).
Bar graphs showing the results of the quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) in the control (ERM cells) and experimental cells. The expression levels of all the stem cell markers, including Nanog, Oct3/4, Sox2, and Klf4 were significantly higher in the DSLCs and iPS cells when compared to those in the controls (**p < 0.01, *p < 0.05; Kruskal–Wallis test, n = 5; a–d). No significant increases in the expression levels of the odontogenic markers, except for Ambn, were observed in the DSLCs (e–g). In the cells treated with 5Aza alone for 1 week and followed by ESCM alone for another week (5Aza2w), the expression levels of Nanog were significantly upregulated (*p < 0.05; a–g). No significant increase in the expression level of any of the markers was noted in the Pro-DSLCs, the cells treated with 5Aza alone for 1 week (5Aza1w), the cells treated with Vpa alone for 1 week (Vpa1w), and the cells treated with Vpa alone for 1 week and followed by ESCM alone for another week (Vpa2w; a–g). No significant increase in the expression level of the tumorigenic marker, Pcna, was observed under any of the culture conditions (h).
The expression levels of NANOG and OCT3/4 were ascertained at the protein level in the Pro-DSLCs and DSLCs via immunocytochemistry; both cells showed positive staining for the two markers. iPS cells were used as positive controls (Fig. 4A). The percentage of SSEA-4-positive cells in the Pro-DSLCs and DSLCSs was significantly higher than that in the controls (ERM cells) (p < 0.01; Fig. 4Ba,b).
Graphs showing the results of the immunofluorescence (A) and flow cytometry (B) tests in the control (ERM cells) and experimental cells. The Pro-DSLCs and DSLCs showed positive staining for NANOG and OCT3/4 (A). Merge, nuclear visualization (blue), NANOG (green), and OCT3/4 (red) visualization. Scale bar = 100 µm. Magnification × 400. The controls, Pro-DSLCs, and DSLCs were stained with SSEA-4 (B). The percentage of SSEA-4-positive cells among the Pro-DSLCs, and DSLCs was significantly higher than that in the controls (**p < 0.01; χ2 tests, n = 5). Gating the SSEA-4-positive cells by flow cytometry (Ba). Analysis of the percentage of SSEA-4-positive/ negative cells (Bb).
Quantitative methylation-specific PCR analysis (qMSP) revealed that the DNA methylation percentage levels of Oct3/4, Sox2, and Klf4 in the cells treated with 5Aza alone (5Aza1w, and 5Aza2w), Pro-DSLCs, and DSLCs were significantly lower than those in the controls (ERM cells) (p < 0.01; Fig. 5A). Furthermore, the cells in each group of Vpa1w, Vpa2w, Pro-DSLCs, DSLCs and the controls (ERM cells) showed increased HDAC activity in a time-dependent manner. The HDAC activities in the Pro-DSLCs, DSLCs, and Vpa1w were significantly lower than those in the controls (ERM cells) (p < 0.01, 0.05; Fig. 5B). In order to verify the reliability of the MSP data, DNA methylation analysis was performed at the CpG sites using the quantitative pyrosequencing analysis (qPSQ). The DNA methylation percentage levels of the CpG sites in Oct3/4 and Sox2 in the cells treated with 5Aza alone (5Aza1w, and 5Aza2w), Pro-DSLCs, and DSLCs were significantly lower than those in the controls (ERM cells) (p < 0.01; Supplementary Fig. S1).
Results of the quantitative methylation-specific PCR (qMSP) (A) and in situ HDAC activity assay (B). DNA methylation of Oct3/4, Sox2, and Klf4 was analyzed using qMSP. The DNA methylation percentage levels of Oct3/4, Sox2, and Klf4 in the cells treated with 5Aza alone (5Aza1w, and 5Aza2w), Pro-DSLCs, and DSLCs were significantly lower than those in the controls (ERM cells) (**p < 0.01; χ2 tests, n = 5). Each sample showed an increase in HDAC activity in a time-dependent manner (B). HDAC activities were significantly lower in the Pro-DSLCs, DSLCs, and the cells treated with Vpa alone for 1 week (Vpa1w) when compared to those in the controls (ERM cells) (**p < 0.01, *p < 0.05; Kruskal–Wallis test, n = 5).
The controls (ERM cells), Pro-DSLCs, and DSLCs were cultured in ECs differentiation medium for 1 week, MSCs differentiation medium for 1 week, and osteogenic differentiation medium for 3 weeks. The mRNA expression levels of the endothelial cell markers platelet and endothelial cell adhesion molecule 1 (Cd31), C34 molecule (Cd34), and cadherin 5 (Cd144), mesenchymal stem cell-negative marker protein tyrosine phosphatase receptor type C (Cd45), mesenchymal stem cell-positive markers Thy-1 cell surface antigen (Cd90), and endoglin (Cd105); osteogenic markers collagen type I alpha 2 chain (Col1a2), osteopontin (Opn), and osteocalcin (Oc), and ameloblastic marker Amel were evaluated. ECs and MSCs were used as positive controls. The expression levels of Cd31 and Cd144 in Pro-DSLCs and ECs, and Cd34 in DSLCs and ECs, cultured in the endothelial differentiation medium were significantly higher than those in the controls (p < 0.01, 0.05; Fig. 6Aa–c). The expression levels of Cd45 in the DSLCs and MSCs cultured in MSCs differentiation culture medium were significantly lower than those in the controls (p < 0.01, 0.05; Fig. 6Ad). The expression levels of Cd90, and Cd105 in the Pro-DSLCs and MSCs were significantly higher than those in the controls (p < 0.01, 0.05; Fig. 6Ae,f).The mRNA expression levels of Col1a2 in Pro-DSLCs and MSCs, Opn in Pro-DSLCs and DSLCs, and Oc in Pro-DSLCs, DSLCs and MSCs, cultured in osteogenic differentiation culture medium were significantly higher than those in the controls (p < 0.01, 0.05; Fig. 6Ba–c); alternatively, a significant decrease in the mRNA levels of Amel were observed in the Pro-DSLCs and DSLCs when compared to those in the controls (p < 0.01; Fig. 6Bd). Flow cytometry revealed a significant increase in the number of cells stained with OC and alkaline phosphatase (ALP) in the Pro-DSLCs and DSLCs when compared to those in the controls (p < 0.01; Fig. 6C). The Pro-DSLCs, DSLC, and MSCs showed increased positive staining for Alizarin Red in a time-dependent manner (Supplementary Fig. S2). The calcification levels determined by Alizarin Red staining in the Pro-DSLCs, DSLCs, and MSCs were significantly higher than those in the controls (p < 0.01, 0.05; Fig. 6D).
Graphs showing the differentiation of controls (ERM cells), Pro-DSLCs, and DSLCs into ECs, MSCs, and osteoblasts. The expression levels of Cd31 and Cd144 (endothelial cell markers) in Pro-DSLCs and ECs, and Cd34 (endothelial cell marker) in DSLCs and ECs cultured in the endothelial differentiation medium were significantly higher than those in the controls (**p < 0.01, *p < 0.05; Kruskal–Wallis test, n = 5; Aa–c). The expression levels of Cd45 (mesenchymal stem cell-negative marker) in the DSLCs and MSCs were significantly lower than those in the controls (**p < 0.01, *p < 0.05; Kruskal–Wallis test, n = 5; Ad). The expression levels of Cd90 and Cd105 (mesenchymal stem cell-positive markers) in the Pro-DSLCs and MSCs were significantly higher than those in the controls (**p < 0.01, *p < 0.05; Ae,f). The mRNA expression levels of osteogenic markers Col1a2 in Pro-DSLCs and MSCs, Opn in Pro-DSLCs and DSLCs, and Oc in Pro-DSLCs, DSLCs and MSCs, cultured in osteogenic differentiation culture medium were significantly higher than those in the controls (**p < 0.01, *p < 0.05; Kruskal–Wallis test, n = 5; Ba–c). Conversely, the significantly decreased mRNA levels of Amel (odontogenic marker) were observed in the Pro-DSLCs and DSLCs compared to those in the controls (**p < 0.01; Bd). The expression of OC and ALP was evaluated using a flow cytometer (C). Flow cytometry revealed a significant increase in the number of cells stained with OC and ALP in the Pro-DSLCs and DSLCs when compared to those in the controls (**p < 0.01; χ2 tests, n = 5). The calcification levels determined by Alizarin Red were significantly higher in the Pro-DSLCs, DSLCs, and MSCs than those in the controls (**p < 0.01, *p < 0.05; Kruskal–Wallis test, n = 5; D).
Discussion
In the present study, epigenetic agents were shown to stimulate the transition of ERM into stem cell-like cells. The expression of stem cell markers was significantly upregulated in ERM cultured in ESCM for an additional week after stimulation with the combination of 5Aza and Vpa (DSLCs). The DSLCs derived from the epithelial cells were able to differentiate into mesenchymal-like cells and exhibit increased expression levels of each of the mesenchymal differentiation markers under different culture conditions. Similarly, the Pro-DSLCs were also differentiated into mesenchymal-like cells under the various culture conditions. These findings indicated that ERM could be stimulated for dedifferentiation into stem cell-like cells via both demethylation and deacetylase inhibition. The differentiation of Pro-DSLCs into mesenchymal-like cells, despite no significant increases in the mRNA expression levels of any of the stem cell markers, may be attributed to direct reprogramming. Positive stainings for NANOG and OCT3/4 were observed in the Pro-DSLCs. The cells that were positively stained with NANOG and OCT3/4 did not present with high mRNAs levels of the corresponding genes as seen in the case of the iPS or stem cells17, which is termed as highly permissive state cells18. Highly permissive cells are generally observed during the process of direct reprogramming. The Pro-DSLCs might represent a type of the highly permissive state cells and play a role in direct reprogramming. The ERM cells were cultured in ESCM for an additional week after the cells were stimulated with the epigenetic agents to generate DSLCs, as described previously8. Pro-DSLCs can be reprogrammed into mesenchymal cells; therefore, culture period in ESCM alone after epigenetic stimulation may not be required for ERM and some other cells to induce direct reprogramming of the cells into other cells. Further investigations are required to confirm this speculation.
Several genes have been epigenetically expressed in iPS cells5. Consequently, epigenetic agents, such as DNMTi and HDACi have been used to generate and improve the production of reprogramming cells6,7,8,9,10. Fibroblasts have been mainly used as an original cell for the generation of iPS and reprogramming cells19. Although the reprogramming efficiency of fibroblasts transducted with Yamanaka factors was improved by the DNMTi and HDACi, the cells could not be completely generated into stem cells without the Yamanaka factors20. The DSLCs in the current study expressed Ambn and other stem cell markers, indicating that these cells were not completely transformed into stem cells, unlike the iPS cells. Thus, the use of epigenetic agents alone may not be sufficient to generate stem cells. In previous studies, fibroblasts were directly reprogrammed into insulin-secreting cells by epigenetic agents in the absence of the Yamanaka factor17,20. Both DSLCs and Pro-DSLCs could be differentiated into other cell types in the present study. The epigenetic agents may be involved in the direct reprogramming of the cells into other cell types via pleiotropic stem-like cells. Both DSLCs and Pro-DSLCs originated from ERM, a type of keratinocyte. The efficiencies of the cells that are transformed into pleiotropic stem-like cells may not matter if they are originally derived from fibroblasts or epithelial cells. The efficiencies of the iPS cells generated from primary keratinocytes have been reported to be less than that of cells derived from fibroblasts19. In another study, the efficiency of reprogramming was higher in immature cells when compared to that in the mature cells21. According to some studies, ERM cells are immature with stem cell-like features and properties15,22. Hence, they may have a higher potential to generate pluripotent stem cells than keratinocytes derived from other types of stratified squamous epithelium. Cell populations contain less than 1% of side population (SP) cells23,24; therefore, the ERM might contain only a small number of SP cells. In fact, the positive staining for both NANOG and OCT3/4 was hardly observed in the ERM (controls). The SP cells derived from tumor cells had highly growth potentials and could be inhibited by either DNMTi or HDACi 25,26. The small number of SP cells might not grow as a part of DSLCs and Pro-DSLCs. The use of specific media for the differentiation of ES or iPS cells into ECs27,28, MSCs29, and osteogenic cells30 have been reported. In the present study, we used the same media as those reported in the afore-mentioned studies, and each one had induced the differentiation of Pro-DSLCs and DSLCs into mesenchymal-like cells. The expression levels for the osteogenic markers in these mesenchymal-like cells were relatively weak as compared to the positive controls. The expression levels of Opn in Pro-DSLCs and DSLC were higher than those in the positive controls, whereas the expression levels of Oc in Pro-DSLCs and DSLC were lower than those in the positive controls. The calcification levels in Pro-DSLCs and DSLC were lower than those in the positive controls. Early mature osteoblasts express Opn, and the expression of Oc was upregulated during osteoblast maturation31. The differences in the expression level of osteogenic markers between the mesenchymal-like cells and the positive controls may depend on the level of osteoblastic differentiation. Further investigations are needed to determine more optimal conditions for their mesenchymal differentiations.
In conclusion, the present study demonstrated that the combination of a demethylating agent and a deacetylated inhibitor induced the dedifferentiation of ERM into stem cell-like cells. The progenitor stem-like cells derived from ERM can be directly reprogrammed into mesenchymal-like cells without dedifferentiation into stem-like cells. Isolated ERM may be used for periodontal regeneration because they can be differentiated into several cells that mainly constitute the periodontal tissues. However, further investigations are needed to prove this hypothesis.
Materials and methods
Figure 1 shows a schematic diagram of the experiment in the current study (Fig. 1).
Culture of epithelial cells from porcine ERM
Epithelial cells were isolated from porcine ERM and cultured using the method described by Brunette et al., and Kurashige et al32,33. Six month-old porcine jaws were obtained from the livestock sales division (HOKUREN Federation of Agricultural Cooperatives, Hokkaido, Japan) and transported to our laboratory on ice. Porcine primary premolars were extracted and explants of the periodontal ligament were cultured in Dulbecco’s modified Eagle medium (Sigma-Aldrich, MO, USA) containing 10% fetal bovine serum (Gibco, Thermo Fisher Scientific, MA, USA) and 2% penicillin–streptomycin (Sigma-Aldrich) at 37 °C in an incubator supplied with 5% CO2. An outgrowth composed of both epithelial-like and fibroblast-like cells was observed. The cells that were more resistant to detachment by 10% dispase (Goudousyusei, Tokyo, Japan) were separated into epithelial cell populations and cultured. The cells that were less resistant to detachment were discarded. The separation procedure was repeated several times to remove any fibroblast cells.
Cell viability
The ERM cells were cultured at a cell density of 0.8 × 106 cells/ml in Primate Embryonic Stem Cell Medium (ESCM, ReproCELL, Kanagawa, Japan) containing 5 ng/ml basic fibroblast growth factor (ReproCELL) and 0.5% penicillin–streptomycin using 60 mm dishes under the following conditions: 0.1, 1 or 10 µM of 5Aza (Tokyo Chemical Industry, Tokyo, Japan); 0.2, 2 or 20 mM of Vpa (FUJIFILM Wako Pure Chemical, Osaka, Japan); and no additions (control) for 1 week. Some of the cells were cultured in ESCM alone for another week because ESCM is known to improve the embryonic stem (ES) property of the cells34,35. The cells were then collected from each culture system using 0.25% Trypsin–EDTA (Gibco, Thermo Fisher Scientific). The dead cells were stained with 0.5% trypan blue and the number of viable cells was counted using a hemocytometer.
Quantitative real-time RT-PCR
We determined the mRNA expression levels of the stem cell markers Nanog, Oct3/4, Sox2, and Klf4, odontogenic markers amelogenin (Amel), Ambn, and keratin19 (Krt19), and tumorigenic marker Pcna in each ERM system treated with 1 µM of 5Aza alone for 1 week (5Aza1w), and followed by ESCM alone for another week (5Aza2w), 2 mM of Vpa alone for 1 week (Vpa1w), and followed by ESCM alone for another week (Vpa2w), and the combination of both 5Aza (1 µM) and Vpa (2 mM) for 1 week (Pro-DSLCs), and followed by ESCM alone for another week (DSLCs; Fig. 1). Human iPS cells (253G1) provided by the RIKEN BRC through the National BioResource Project of the MEXT/AMED (Ibaraki, Japan) were used as positive controls for the stem cell markers36. Total RNA was extracted from the cells via the acid guanidine thiocyanate/phenol–chloroform method using TRizol (Invitrogen, Thermo Fisher Scientific MA, USA). One microgram of the RNA sample was reverse-transcribed (SuperScript reverse transcriptase, Invitrogen) according to the manufacturer’s instructions using oligo (dT) 12–18 primers (Invitrogen, Thermo Fisher Scientific). For quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR), a reaction mixture was prepared using the KAPA SYBR Fast qPCR Kit (Kapa Biosystems, Roche, Basel, Switzerland), primers, and RT products. The primer sequences used in this study are shown in Table 1. The qRT-PCR was performed using Light Cycler Nano (Roche). The qPCR conditions included an initial incubation at 50 °C for 2 min, denaturation at 95 °C for 10 min, 45 cycles of denaturation at 95 °C for 15 s, and annealing at 55 °C for 1 min. The relative expression level of each mRNA was calculated using the ΔΔCq method37. Data are shown as a ratio of the target mRNA to the glyceraldehyde-3-phosphate dehydrogenase (Gapdh) mRNA.
Immunostaining
The controls, Pro-DSLCs, and DSLCs were fixed with 60% Acetone/ 40% methanol in a chamber slide (Lab-Tek Chamber Slide System, Nunc, Thermo Fisher Scientific) at room temperature for 15 min. The fixed cells were rinsed with PBS, three times at 10 min intervals. After pre-incubation in blocking buffer (5% normal goat serum, Dako, Glostrup, Denmark) at room temperature for 1 h, the cells were incubated overnight with the primary anti-human antibodies NANOG and OCT3/4 (1: 100; Human Pluripotent Stem Cell 3-Color Immunocytochemistry Kit, R&D Systems, MN, USA) at 4 °C, according to the manufacturer's instructions. The next day, the cells were rinsed alternately with PBST and PBS (3 times at 10 min intervals) and incubated with goat anti-rabbit IgG (H + L) Secondary Antibody Alexa Fluor 488 conjugate (1: 1000; Thermo Fisher Scientific) in the dark at room temperature for 1 h. After the cells were rinsed alternately with PBST and PBS (4 times at 10 min intervals), they were incubated with DAPI Fluoromount-G (SouthernBiotech, AL, USA) for nuclear visualization. iPS cells were used as positive controls for the NANOG and OCT3/4. The cells were observed using a confocal microscope (Nikon EZ-C1, Tokyo, Japan).
Flow cytometry
Flow cytometry was used to analyze the cells stained with stage-specific embryonic antigen-4 (SSEA-4). The controls, Pro-DSLCs, and DSLCs were harvested from each cell system and filtered using a FALCON Cell Strainer (100 μm; Corning, NY, USA). The cells were incubated in the dark for 30 min at 4 °C with SSEA-4 Alexa Fluor 647 antibodies (Human Induced Pluripotent Stem Cell Analysis and Sorting Kit, BD Biosciences, NJ, USA), and further incubated for 10 min at 4 °C with 7-Amino-Actinomycin D (7-AAD, BD Biosciences). SSEA-4 was analyzed using a flow cytometer (BD FACSAriaIIIu, BD Biosciences).
Quantitative methylation-specific PCR analysis
In order to confirm the DNA methylation levels of the cells in the experiments, genomic DNA was extracted from each cell culture system using the DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany). The DNA sample (500 ng) was treated with sodium bisulfite using the EpiTect Fast Bisulfite Kits (Qiagen). The DNA methylation of Oct3/4, Sox2, and Klf4 was analyzed via SYBR green-based qMSP after checking for the presence of CpG islands around the promoter region in the UCSC Genome Browser (http://genome.ucsc.edu/index.html). Two sets of qMSP primers were designed using MethPrimer (http://www.urogene.org/methprimer/; Supplementary Figs. S3A, B)38; one for unmethylated and the other for methylated DNA sequences (Table 1). For the qMSP, the bisulfite-treated DNA template was mixed with KAPA SYBR Fast qPCR Kit and a pair of primers. The qMSP conditions included an initial incubation at 50 °C for 2 min, denaturation at 95 °C for 10 min, 50 cycles of denaturation at 95 °C for 15 s, and annealing at 55 °C for 1 min. The percentage of DNA methylation in the samples was estimated using the following equation:
where M is the copy number of methylated DNA, U is the copy number of unmethylated DNA, and ΔCq = CqU − CqM39.
Quantitative pyrosequencing analysis
In order to verify the reliability of the MSP data, DNA methylation analysis was performed at the CpG site using the qPSQ method (ALLIANCE Biosystems, Osaka, Japan). The target region of Oct3/4, which contained five CpG sites, and of Sox2, which contained four CpG sites (sequence 1) and three CpG sites (sequence 2), were amplified using the forward primer and biotinylated reverse primer (PSQ Assay Design; Qiagen, Hilden, Germany; Table 2; Supplementary Fig. S3C). However, it was not possible to perform the analysis due to an error in the PCR primer design owing to the dense CG region in Klf4. Genomic DNA (200 ng) was modified by sodium bisulfite using the EZ DNA Methylation-Lighting kit (Zymo Research, CA, USA), according to the manufacturer’s instructions. Bisulfite-modified DNA was amplified in a 20-μl reaction volume containing the PCR pre-mixture (Enzynomics, Korea), 1 μl of Primer-S (10 pmol/μl), and 1 μl of biotinylated-Primer-As (10 pmol/μl). The PCR products were visualized on 2% agarose gel via ethidium bromide staining for verification.
A DNA template was prepared from 16–18 μl of the biotinylated PCR product using streptavidin Sepharose HP beads (Cytiva, GE Healthcare Life Sciences, MA, USA) and multichannel pipets according to the PSQ 96 sample preparation guide. Fifteen picomoles of the respective sequencing primer were added for analysis. The sequencing was performed on a PyroMark ID system (Qiagen) using the Pyro Gold reagents kit (Qiagen), according to the manufacturer’s instruction and without any further optimization. The methylation index (MtI) of each target region in each sample was calculated as the average value of mC/(mC + C) for all the examined CpGs. All experiments included samples without templates as negative control.
In situ HDAC activity assay
The HDAC activity of the cells in the experiments was confirmed with the In Situ HDAC Activity Fluorometric Assay Kit (BioVision Mountainview, CA, USA). Fluorescence signals were detected following excitation at 380 nm and emission at 450 nm using a fluorescence microplate reader (Infinite F200, TECAN, Männedorf, Switzerland).
Differentiation of the cells into endothelial cells, mesenchymal stem cells, and osteoblasts
The controls (ERM cells), Pro-DSLCs, and DSLCs were cultured (0.8 × 106 cells/ml) with the EGM-2 BulletKit (Lonza, Verviers, Belgium) for ECs differentiation (1 week), MSCGM BulletKit (Lonza) for MSCs differentiation (1 week), and RPMI 1640 Medium (Gibco, Thermo Fisher Scientific) with Osteoblast-Inducer Reagent (Takara Bio, Shiga, Japan) for osteogenic differentiation (3 weeks) at 37 °C in an incubator supplied with 5% CO2. The mRNA expression levels of the endothelial cell markers Cd31, Cd34, and Cd144, mesenchymal stem cell-negative marker Cd45, mesenchymal stem cell-positive markers Cd90, and Cd105; osteogenic markers Col1a2, Opn, and Oc, and ameloblastic marker Amel were evaluated. ECs (Cosmo Bio, Tokyo, Japan) and MSCs (Cell Biologics, IL, USA) were used as positive controls.
The expression levels of the protein markers for osteogenic differentiation were also confirmed. The controls (ERM cells), Pro-DSLCs, and DSLCs were cultured in RPMI 1640 Medium with Osteoblast-Inducer Reagent for 3 weeks and filtered using a FALCON Cell Strainer (100 μm; Corning). The cells were incubated in the dark for 30 min at 4 °C with Alexa Fluor 488 mouse anti-Human alkaline phosphatase (BD Biosciences) and PE mouse anti-human osteocalcin (BD Biosciences), followed by staining with 7-AAD (BD Biosciences). Alkaline phosphatase (ALP) and OC staining in the cells were analyzed by flow cytometry (BD FACSAriaIIIu, BD Biosciences). Furthermore, the controls (ERM cells), Pro-DSLCs, DSLCs, and MSCs were cultured in RPMI 1640 Medium with Osteoblast-Inducer Reagent in a 24-well plate (IWAKI, Tokyo, Japan) for 3 weeks. Subsequently, the calcification levels were examined using the Alizarin Red staining set (PG Research, Tokyo, Japan), and absorbance at 450 nm was determined using a microplate reader.
Statistical analysis
The experiments were performed in quintuplicate, except for qPSQ (quadruplicate). Data were analyzed using Kruskal–Wallis or χ2 tests, with a p < 0.01 or < 0.05 considered as significant. The data are presented as means ± standard error. All statistical analyses were performed using SPSS version 23 (IBM, NY, USA).
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Yasui, T. et al. Isolation of dental pulp stem cells with high osteogenic potential. Inflamm. Regen. 37, 8 (2017).
Tamaoki, N. et al. Dental pulp cells for induced pluripotent stem cell banking. J. Dent. Res. 89, 773–778 (2010).
Yan, X. et al. iPS cells reprogrammed from human mesenchymal-like stem/progenitor cells of dental tissue origin. Stem Cells Dev. 19, 469–480 (2010).
Malhotra, N. Induced pluripotent stem (iPS) cells in dentistry: a review. Int. J. Stem Cells 9, 176–185 (2016).
Takahashi, K. & Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006).
Mikkelsen, T. S. et al. Dissecting direct reprogramming through integrative genomic analysis. Nature 454, 49–55 (2008).
Huangfu, D. et al. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat. Biotechnol. 26, 1269–1275 (2008).
Huangfu, D. et al. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat. Biotechnol. 26, 795–797 (2008).
Li, Y. et al. Generation of iPSCs from mouse fibroblasts with a single gene, Oct4, and small molecules. Cell Res. 21, 196–204 (2011).
Li, X. et al. Generation of neural progenitors from induced Bama miniature pig pluripotent cells. Reproduction 147, 65–72 (2013).
Duncan, H. F., Smith, A. J., Fleming, G. J. & Cooper, P. R. Epigenetic modulation of dental pulp stem cells: implications for regenerative endodontics. Int. Endod. J. 49, 431–446 (2016).
Peng, Z., Liu, L., Wei, X. & Ling, J. Expression of Oct-4, SOX-2, and MYC in dental papilla cells and dental follicle cells during in-vivo tooth development and in-vitro co-culture. Eur. J. Oral Sci. 122, 251–258 (2014).
Juuri, E., Isaksson, S., Jussila, M., Heikinheimo, K. & Thesleff, I. Expression of the stem cell marker, SOX2, in ameloblastoma and dental epithelium. Eur. J. Oral. Sci. 121, 509–516 (2013).
Xiong, J., Mrozik, K., Gronthos, S. & Bartold, P. M. Epithelial cell rests of Malassez contain unique stem cell populations capable of undergoing epithelial-mesenchymal transition. Stem Cells Dev. 21, 2012–2025 (2012).
Tsunematsu, T. et al. Human odontogenic epithelial cells derived from epithelial rests of Malassez possess stem cell properties. Lab. Invest. 96, 1063–1075 (2016).
Rincon, J. C., Young, W. G. & Bartold, P. M. The epithelial cell rests of Malassez–a role in periodontal regeneration?. J. Periodontal. Res. 41, 245–252 (2006).
Pennarossa, G. et al. Brief demethylation step allows the conversion of adult human skin fibroblasts into insulin-secreting cells. Proc. Natl. Acad. Sci. USA 110, 8948–8953 (2013).
Brevini, T. A. et al. Phenotype switching through epigenetic conversion. Reprod. Fertil. 27, 776–783 (2015).
Stadtfeld, M. & Hochedlinger, K. Induced pluripotency: history, mechanisms, and applications. Genes. Dev. 24, 2239–2263 (2010).
Pennarossa, G. et al. Reprogramming of pig dermal fibroblast into insulin secreting cells by a brief exposure to 5-aza-cytidine. Stem Cell Rev. 10, 31–43 (2014).
Takahashi, K. & Yamanaka, S. Induced pluripotent stem cells in medicine and biology. Development 140, 2457–2461 (2013).
Padma Priya, S. et al. Odontogenic epithelial stem cells: hidden sources. Lab. Invest. 95, 1344–1352 (2015).
Iohara, K. et al. Side population cells isolated from porcine dental pulp tissue with self-renewal and multipotency for dentinogenesis, chondrogenesis, adipogenesis, and neurogenesis. Stem Cells 24, 2493–2503 (2006).
Kato, K. Stem cells in human normal endometrium and endometrial cancer cells: characterization of side population cells. Kaohsiung J. Med. Sci. 28, 63–71 (2012).
Kato, K. et al. Sodium butyrate inhibits the self-renewal capacity of endometrial tumor side-population cells by inducing a DNA damage response. Mol. Cancer Ther. 10, 1430–1439 (2011).
Kim, N. Y., Kim, M. C. & Kim, Y. Hypomethylation reduced the aggressive potential of human malignant mesothelioma cells. Cancer Gene Ther. 23, 425–432 (2016).
Kim, G. D. et al. Differentiation of endothelial cells derived from mouse embryoid bodies: a possible in vitro vasculogenesis model. Toxicol. Lett. 180, 166–173 (2008).
Tan, K. S. et al. Molecular pathways governing development of vascular endothelial cells from ES/iPS cells. Stem Cell Rev. 9, 586–598 (2013).
Villa-Diaz, L. G. et al. Derivation of mesenchymal stem cells from human induced pluripotent stem cells cultured on synthetic substrates. Stem Cells 30, 1174–1181 (2012).
Tokita, R. et al. Differentiation behavior of iPS cells cultured on PLGA with osteoinduction medium. Dent. Mater. J. 36, 103–110 (2017).
Komori, T. Regulation of bone development and maintenance by Runx2. Front. Biosci. 13, 898–903 (2008).
Brunette, D. M., Melcher, A. H. & Moe, H. K. Culture and origin of epithelium-like and fibroblast-like cells from porcine periodontal ligament explants and cell suspensions. Arch Oral Biol 21, 393–400 (1976).
Kurashige, Y. et al. Profiling of differentially expressed genes in porcine epithelial cells derived from periodontal ligament and gingiva by DNA microarray. Arch. Oral Biol. 53, 437–442 (2008).
Nakatsuji, N. & Suemori, H. Embryonic stem cell lines of nonhuman primates. Sci. World J. 2, 1762–1773 (2002).
Garcia-Gonzalo, F. R. & Izpisúa Belmonte, J. C. Albumin-associated lipids regulate human embryonic stem cell self-renewal. PLoS ONE 3, e1384 (2008).
Nakagawa, M. et al. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat. Biotechnol. 26, 101–106 (2008).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408 (2001).
Li, L. C. & Dahiya, R. MethPrimer: designing primers for methylation PCRs. Bioinformatics 18, 1427–1431 (2002).
Lu, L., Katsaros, D., de la Longrais, I. A., Sochirca, O. & Yu, H. Hypermethylation of let-7a-3 in epithelial ovarian cancer is associated with low insulin-like growth factor-II expression and favorable prognosis. Cancer Res. 67, 10117–10122 (2007).
Acknowledgements
This research was supported by the Grant-in-Aid for Early-Career Scientists (B) from the Japan Society for the Promotion of Science (No. 18K17091).
Author information
Authors and Affiliations
Contributions
K.Y. and Y.A. have designed the study. K.Y., O.U., Y.K., D.P., A.O., P.N., D.H., T.M., F.H., R.T. and J.S. have been involved in data collection and data analysis. K.Y., O.U., Y.K. and M.S. have been involved in data interpretation. K.Y., D.P. and Y.A. have been involved in drafting the manuscript and revising it critically and have given final approval of the version to be published.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yoshida, K., Uehara, O., Kurashige, Y. et al. Direct reprogramming of epithelial cell rests of malassez into mesenchymal-like cells by epigenetic agents. Sci Rep 11, 1852 (2021). https://doi.org/10.1038/s41598-020-79426-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-79426-4
This article is cited by
-
Analysis of the cells isolated from epithelial cell rests of Malassez through single-cell limiting dilution
Scientific Reports (2022)
-
Increased integrity of cell–cell junctions accompanied by increased expression of claudin 4 in keratinocytes stimulated with vitamin D3
Medical Molecular Morphology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.