Abstract
Analysis of large gene expression datasets from biopsies of cancer patients can identify co-expression signatures representing particular biomolecular events in cancer. Some of these signatures involve genomically co-localized genes resulting from the presence of copy number alterations (CNAs), for which analysis of the expression of the underlying genes provides valuable information about their combined role as oncogenes or tumor suppressor genes. Here we focus on the discovery and interpretation of such signatures that are present in multiple cancer types due to driver amplifications and deletions in particular regions of the genome after doing a comprehensive analysis combining both gene expression and CNA data from The Cancer Genome Atlas.
Similar content being viewed by others
Introduction
Gene co-expression signatures in cancer often involve genomically co-localized genes resulting from the presence of various biological mechanisms that include, but are not limited to, the copy number alterations (CNAs) of malignant cells1 and the immune response against cancer cells2. For example, ERBB2, GRB7, MIEN1 are among the genes co-expressed in breast cancer due to the HER2 amplicon3, while HLA-DPA1, HLA-DPB1, HLA-DRA are among the genes co-expressed in the MHC Class II immune cluster4. Any co-expression signatures that are consistently present in many different cancer types are referred to as "pan-cancer" signatures, representing universal (tissue-independent) biomolecular events in cancer5,6,7,8. Ref.8 studied co-expressed genes in immune cells. There are several techniques for identifying co-expression signatures involving genomically co-localized genes9,10,11.
We have proposed an unsupervised algorithm to identify genome-wide co-expression signatures known as attractor metagenes5, a version of which was focusing on genomically co-localized signature finding. Attractor metagenes have been used successfully for cancer biomarker discovery12,13,14.
The identification of genomically co-localized gene signatures can shed light on some complex cancer-related biological mechanisms, especially the tumor driving events caused by CNAs. CNAs involve amplified or deleted DNA regions, which have been generated by the chromosomal instability of malignant cells. If such CNAs are frequently present in cancer cells contained in multiple cancer samples, this suggests that they have an evolutionary advantage and therefore are "driver" CNAs with the tendency to create subclones in the heterogeneous tumors. Although CNAs may include in some cases a single or few oncogenes or tumor suppressor genes, in which case their pan-cancer identification covers a small genomic region containing that gene15,16, they typically influence DNA regions covering many genes, implying that some of these genes have synergistic functions in tumorigenesis17. Here we focus on pan-cancer CNAs containing multiple genes.
The previous work on identifying pan-cancer CNAs18,19 only made use of data resulting from analysis of the genomes in the malignant cells. However, the evolutionary advantage of CNAs is based on the expression of particular genes located within the CNA genomic region. Therefore, analysis of gene expression data provides additional valuable information20,21,22,23,24. Some among the list of the consistently co-expressed genes, including the first and the last when sorted in terms of their genomic location, play some role in tumorigenesis and it is possible that this role is due to their synergistic functions. More generally, gene expression analysis provides helpful information for the identification of the driver genes in each preserved CNA genomic region by pointing to those genes that are consistently amplified or deleted.
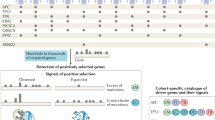
In this paper, we use a novel methodology for the identification of pan-cancer co-localized gene signatures containing no less than five strongly co-expressed genes that are due to CNAs, by making use of gene expression as well as CNA data from The Cancer Genome Atlas (TCGA). Part of this method applies a pan-cancer version of a genomically co-localized attractor algorithm, which is an extension of our previous work5. Our work identified several pan-cancer CNAs not previously detected in the pan-cancer analysis of CNAs18,19, such as 1q41, 7p22.3, 8q13.1-24.3, 10p12.1, 19q13.12, 20p13 (amplifications) and 1p36.33-36.22, 16q22.1 and 17p13.2 (deletions).
Results
Summary
We applied the pan-cancer genomically co-localized attractor algorithm (Materials and Methods) to the TCGA expression data of 56,830 genes and 8593 tumor cases covering eighteen major types of cancer (Table S1), using a window size of 150 genes. This resulted in the identification of 101 pan-cancer genomically co-localized gene signatures (Table S2). To designate such signatures as being caused by driver CNAs, we reasoned that they should satisfy two conditions simultaneously: They should exhibit a high association between their corresponding levels of gene expression and CNA values, and at the same time their genomic regions should frequently appear as CNAs in multiple cancer types. 76 signatures had high expression/CNA level association (P < 0.05, Table S3, Materials and Methods). 54 signatures had high amplification or deletion frequency (Table S4, Materials and Methods). 37 genomically co-localized signatures satisfied both conditions above, and were designated as being caused by CNAs in cancer cells containing cooperative oncogenes/tumor suppressor genes (Tables S3 and S4). Among those 37 genomically co-localized signatures, 25 signatures correspond to pan-cancer amplifications (Table 1 and Fig. 1) and 12 signatures correspond to pan-cancer deletions (Table 2 and Fig. 2). Figures 1 and 2 include bars whose height is proportional to the weight of each gene in the co-expression signature (Materials and Methods). References confirming the oncogenic roles of amplified gene signatures and the tumor suppressing roles of the deleted gene signatures are listed in Tables 1 and 2.
Some of the identified signatures are located genomically close to each other. This suggests that each of them, by itself, has sufficient evolutionary advantage (indeed, we observed that the expression levels of adjacent genomically co-localized signatures are often independent of each other), but it is also possible for an amplicon to cover multiple such regions simultaneously (Materials and Methods, Table S5).
To provide insights of the underlying biological significance in particular examples, we analyze some of such CNAs in the following sections.
Genomically co-localized signatures associated with 1q21.3-q41 amplification
We identified signature VPS72 and signature FLAD1 located on 1q21.3 amplicon (Fig. 1A,B). The expression level of signature VPS72 is strongly associated with the expression level of signature FLAD1 (Figure S1) and these two signatures have a co-amplification frequency of 91.6% (Figure S2, Table S5). We also identified another genomically co-localized signature, RAB3GAP2, located on 1q41, which has not been detected as a pan-cancer amplicon18,19. We observed that the expression level of signature FLAD1 is not associated with the expression level of signature RAB3GAP2 (Figure S3), although they are co-amplified in 79.8% of the cancer cases (Figure S4, Table S5).
GSEA86 (Gene Set Enrichment Analysis) was applied to the genes of the three signatures VPS72, FLAD1 and RAB3GAP2, concluding that these genes are enriched with the GO (Gene Ontology) term ‘Mitochondrion’ (P < 10−7, Q < 10−3), thus potentially helping the efforts to shed light on the underlying biological mechanism.
Genomically co-localized signatures associated with 8q13.1–24.3 amplification
We identified three genomically co-localized signatures located on the 8q arm: ARMC1, UTP23 and SHARPIN (Fig. 1L,M,N). The expression plots between signature ARMC1 and signature UTP23 show that they are associated with each other (Figure S5), and that they are co-amplified in 76.2% of the cancer cases (Figure S6, Table S5). This suggests that there is a synergistic effect between them. On the other hand, the expression levels of signature UTP23 and signature SHARPIN are independent (Figure S7) although these two signatures are co-amplified in 77.6% of the cancer cases (Figure S8, Table S5).
Genomically co-localized signatures associated with 1p36.33–22 deletion
We identified two genomically co-localized signatures, UBE2J2 and MIIP (Fig. 2A,B) located on 1p36.33-36.22 that have not been detected as pan-cancer deleted regions18,19. The expression levels of signature UBE2J2 and signature MIIP are strongly associated with each other (Figure S9) with co-deletion frequency of 70.3% (Figure S10, Table S5), suggesting these GLAs can either independently exist or be co-deleted. Among the genes in these two signatures, gene AURKAIP1 down-regulates the Aurora-A oncogene67. Gene FAAP20 is needed in DNA repair pathway68. The deletion of gene MIIP can induce chromosomal instability69. Tumor suppressor gene MAD2L2 inhibits cancer growth70. GSEA was applied to the genes of the two signatures UBE2J2 and signature MIIP, concluding that these genes are enriched in the GO term ‘Negative Regulation of Cellular Component Organization’ (P < 10−4, Q < 0.05), suggesting potential mechanisms associated with the evolutionary advantage of their simultaneous deletion.
Comparison with previous TCGA studies
We compared our results with the tumor driving CNAs detected in Refs.18,19. On the one hand, several CNAs that we identified by our joint expression/CNA analysis were missed in both of those references. On the other hand, because our algorithm was designed to detect at least five consistently strongly co-expressed genes (Materials and Methods), we do not include the “peak CNAs”, as well as those CNVs containing less than five co-expressed genes (Table S6), which were obtained in Refs.18,19. Such peak CNAs include those containing MYC, CCND1, METTL1, NKX2-1, EGFR, FGFR1, KRAS, CCNE1, CRKL, CDKN2A, FHIT, WWOX, PTPRD, MACROD2, PRKN, LRP1B, RNA5SP174, PLK2, and RBFOX1 (Figures S11, S12, Table S6). Despite the small number of potential driver genes in peak CNAs, our algorithm can help identify the cooperative effects between those genes. For example, signature MYC (Figure S11A) consists of genes in the neighborhood of gene MYC. Among them, the long non-coding RNA PVT-1 has the second strongest association with the signature, suggesting that PVT-1 also plays a role in tumorigenesis, consistent with the previous conclusion87 that PVT-1 and MYC have cooperative effect in cancer. Furthermore, FAM84B, another gene adjacent to MYC, is the fifth top-ranked gene associated with the signature, consistent with its identified role88 of strengthening the function of MYC. Examples of signatures containing less than five genes are those containing ATAD1 and PTEN in 10q23.31 (Table S6), THAP3, 2BTB48, PARK7 in 1p36.31 (Table S6), and STK25, ATG4B, ING5, THAP4 in 2q37.3 (Table S6). All signatures identified on the CNAs listed in Refs.18,19 can be found in Table S6.
Discussion and conclusion
This paper focuses on detecting pan-cancer genomically co-localized gene co-expression signatures associated with amplicons or deleted regions, identifying several novel pan-cancer CNAs. Such signatures contain oncogenes or tumor suppressor genes and result from the cooperative effect of some of their member genes. We have also found that some amplified regions contain multiple genomically co-localized signatures with different tumorigenesis functions, which are occasionally amplified separately. Previous studies (Refs.20,24) used the association between expression and CNA levels as part of their methods to determine whether a gene is likely to be an oncogene or a tumor suppressor gene. Therefore, many of such previously identified genes are included in our identified genomically co-localized signatures. For example, gene VPS72 and gene PSMD4 are identified as two oncogenes in Ref.20, and these two genes are identified as cooperative oncogenes co-expressed in signature VPS72. Gene MED21 and gene CCDC91, two oncogenes independently identified in Ref.20, are co-expressed in signature MED21. Genes SYNCRIP and MAP3K7, two tumor suppressor genes reported in Ref.24, are identified as components of signature SYNCRIP in this paper. Similarly, tumor suppressor signature CCAR contains three co-expressed tumor suppressor genes, CHMP7, CCDC25, and INTS9, which were identified as independent tumor suppressor genes in Ref.24. Our analysis not only indicates that genes may be oncogenes or tumor suppressor genes, but also suggests that the co-expressed genes in a genomically co-localized signature have cooperative effects in tumorigenesis due to their simultaneous amplification or deletion.
Materials and methods
Data preparation
We downloaded harmonized TCGA gene expression data processed by HTSeq-FPKM (High-Throughput Seq-Fragments Per Kilobase of transcript per Million mapped reads) workflow and copy number segment (CNS) data generated by Affymetrix SNP 6.0 platform from Genomic Data Commons89 using the TCGAbiolinks package from Bioconductor. We also used the PanCancer Atlas Clinical Data Resource (CDR) Outcome from https://gdc.cancer.gov/about-data/publications/pancanatlas.
We focused on eighteen major types of cancer: bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), colon adenocarcinoma (COAD), head and neck squamous cell carcinoma (HNSC), kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), brain lower grade glioma (LGG), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), ovarian serous cystadenocarcinoma (OV), prostate adenocarcinoma (PRAD), sarcoma (SARC), skin cutaneous melanoma (SKCM), stomach adenocarcinoma (STAD), thyroid carcinoma (THCA), uterine corpus endometrial carcinoma (UCEC), covering 8593 cancer cases. The number of cases in each type of cancer is given in Table S1.
The log2(1 + X) transformed expression data were normalized using the quantile normalization methods implemented in the limma package from Bioconductor. Genes having zero value across all samples from any type of cancer were excluded from the whole datasets. Gene-level CNA values were inferred from their corresponding CNS data. The CNS data are in the form of log-2-ratio, i.e. zero means a normal diploid number of 2, a positive number means amplification, and a negative number represents deletion. If a gene did not fall into any segment in the CNS data, then its CNA value was inferred by the mean value of its two adjacent segments. Each row of an expression/CNA matrix corresponds to a gene (or a signature), while each column corresponds to a cancer case.
Association measurement
The association measure of mutual information (MI) \(I\left( {A;B} \right)\) between two random variables \(A\) and \(B\) is defined by the expected value of \( - {\log}\left( {p_{A} p_{B} /p_{AB} } \right)\), where \(p_{A}\) and \(p_{B}\) are the marginal distributions and \(p_{AB}\) is the joint probability density. We use a spline-based estimator with six bins in each dimension to estimate the MI90 given the two vectors representing the variables. We normalize this estimate by dividing by the maximum of the estimated \(I\left( {A;A} \right)\) and \( I\left( {B;B} \right)\), so that the result has a maximum value of 1 representing complete corlation beeen two variables, and a minimum value of zero representing independence between two variables. We multly by − 1 whenever the Pearson correlation between A and B is negative, so the final association measure can take values between − 1 and 1.
If variables \(A\) and \(B\) both exist in all types of cancer, then the pan-cancer association between \(A\) and \(B\) is defined by the weighted median of the normalized MIs between \(A\) and \(B\) across all types of cancer, where the weights are given by the proportion of samples in each cancer type. Specifically, by using the weighted median, in which the weights are given by the proportion of samples in each cancer type, the evaluation of the pan-cancer association is balanced, ensuring that all samples are treated equally.
Genomically co-localized signature finding algorithm
We first sort all N genes (N = 56,830) based on genomic mid-point and apply a sliding-window preprocessing approach to identify the co-exprsion signatures, as follows. We use each of the N genes as a seed gene, applying the iterative attractor metagene iterative algorithm5, considering only the nearest \(S\) genes (S/2 at each side, or as many as available at chromosomal ends) of this gene according to the genomic sorting (setting window parameter \(S\) = 150 genes, exponent parameter α = 2 and convergence parameter ε = 10–7). The resulting attractor metagene is defined by a weighted average of the expression values of these S + 1 genes. There are S + 1 such weights. The name of the gene with the highest weight is used as the name of this metagene, and the remaining S genes are sorted in terms of their corresponding weights. The strength of each attractor metagene is defined as the fifth highest weight. We filter out metagenes with strength less than 0.5. Therefore, each metagene contains at least five strongly co-expressed genes. The chromosomal range of each metagene is defined by its member genes with weight larger than 0.5. Attractor metagenes with overlapped chromosomal ranges are then merged into one cluster, resulting in a total of L clusters, each of which defines a chromosomal range.
For each of these resulting L chromosomal ranges, we run the attractor metagene algorithm again, using each of the member genes as a seed within the range. If a chromosomal range yields multiple different attractor metagenes, we select the one with the highest strength to represent the chromosomal range. In the end, we generate \(L\) attractor metagenes. We further filter out any attractor metagenes whose top five genes have zero expression values in more than half of the samples. Finally, we filter out the gender-based attractor metagenes located on chromosome X and Y.
Association between the expression levels and the CNA levels of a signature
We use the average of expression/CNA levels of the top five genes of a genomically co-localized signature as a measure of the overall expression/CNA level of this signature. Then the pan-cancer association between the expression levels and CNA levels of a signature (pan-cancer expression-CNA association) is given by the weighted median of the corresponding normalized MIs, where the weights are given by the proportion of samples in each cancer type. We run 10,000 permutations and a random distribution between the permuted expression level and CNA level of each signature in each type of cancer is generated. For each signature, its pan-cancer distribution is obtained by the weighted median of its sorted distribution in each type of cancer. The P value of the pan-cancer expression/CNA association is given by the proportion of the pan-cancer distribution larger than the pan-cancer expression/CNA association and later adjusted using Bonferroni correction. We assume that P = 0.05 defines the threshold of statistical significance.
Signatures located on amplicons or deleted regions across multiple types of cancer
We set thresholds \(t_{{{\text{amp}}}}\) and \(t_{{{\text{del}}}}\) to identify genomically co-localized signatures located on amplified or deleted regions, to be selected so that genes with CNA values larger than \(t_{{{\text{amp}}}}\) are amplified and genes with CNA levels smaller than \(t_{{{\text{del}}}}\) are deleted. The thresholds \(t_{{{\text{amp}}}}\) and \(t_{{{\text{del}}}}\) are set using the empirical distribution of CNS levels in normal samples. vels of a normal sample are first subtracted by the mean CNS value of this sample. Then, for each cancer type \(c\), we obtain \(t_{{{\text{amp}}|c}}\) (\(t_{{{\text{del}}|c}}\)) using the mean value of the top (bottom) 10 percentile CNA values from all the samples in this cancer type. The thresholds \(t_{{{\text{amp}}}}\) (\(t_{{{\text{del}}}}\)) are calculated by the weighted median of \(t_{{{\text{amp}}|c}}\) (\(t_{{{\text{del}}|c}}\)) across the eighteen types of cancer. This gives \(t_{{{\text{amp}}}} = 0.45\) and \(t_{{{\text{del}}}} = - 0.62\).
The amplification and deletion frequencies of each genomically co-localized signature are calculated in each of the eighteen types of cancer. A signature is classified as amplified (deleted) in one type of cancer if its amplification (deletion) frequency is larger than \(t_{{{\text{freq}}}}\), which is empirically set to 3%. We assume that if a signature is amplified (deleted) in more than 6 types of cancer, then this signature is located on a pan-cancer amplicon (deleted region) and assume two adjacent signatures are co-amplified or co-deleted if they have a CNA difference less than 0.1.
References
Fehrmann, R. S. N. et al. Gene expression analysis identifies global gene dosage sensitivity in cancer. Nat. Genet. 47, 115 (2015).
Chifman, J., Pullikuth, A., Chou, J. W., Bedognetti, D. & Miller, L. D. Conservation of immune gene signatures in solid tumors and prognostic implications. BMC Cancer 16, 911 (2016).
Katz, E. et al. A gene on the HER2 amplicon, C35, is an oncogene in breast cancer whose actions are prevented by inhibition of Syk. Br. J. Cancer 103, 401 (2010).
Ting, J.P.-Y. & Trowsdale, J. Genetic control of MHC class II expression. Cell 109, S21–S33 (2002).
Cheng, W.-Y., Yang, T.-H.O. & Anastassiou, D. Biomolecular events in cancer revealed by attractor metagenes. PLOS Comput. Biol. 9, e1002920 (2013).
Ficklin, S. P. et al. Discovering condition-specific gene co-expression patterns using gaussian mixture models: a cancer case study. Sci. Rep. 7, 8617 (2017).
Yang, Y. et al. Gene co-expression network analysis reveals common system-level properties of prognostic genes across cancer types. Nat. Commun. 5, 3231 (2014).
Wang, P. et al. ImmuCo: a database of gene co-expression in immune cells. Nucleic Acids Res. 43, D1133–D1139 (2014).
Dottorini, T. et al. CluGene: a bioinformatics framework for the identification of co-localized, co-expressed and co-regulated genes aimed at the investigation of transcriptional regulatory networks from high-throughput expression data. PLoS ONE 8, e66196 (2013).
Delatola, E. I. et al. SegCorr a statistical procedure for the detection of genomic regions of correlated expression. BMC Bioinform. 18, 333 (2017).
Cai, L. et al. Genomic regression analysis of coordinated expression. Nat. Commun. 8, 2187 (2017).
Ou Yang, T.-H., Cheng, W.-Y., Zheng, T., Maurer, M. A. & Anastassiou, D. Breast cancer prognostic biomarker using attractor metagenes and the FGD3–SUSD3 metagene. Cancer Epidemiol. Biomark. Prevent. 23, 2850 (2014).
Cheng, W.-Y., Yang, T.-H.O. & Anastassiou, D. Development of a prognostic model for breast cancer survival in an open challenge environment. Sci. Transl. Med. 5, 181ra150 (2013).
McCarthy, N. Rising to the challenge. Nat. Rev. Cancer 13, 378–378 (2013).
Beroukhim, R. et al. Assessing the significance of chromosomal aberrations in cancer: Methodology and application to glioma. Proc. Natl. Acad. Sci. 104, 20007–20012 (2007).
Mermel, C. H. et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 12, R41 (2011).
Taylor, A. M. et al. Genomic and Functional approaches to understanding cancer aneuploidy. Cancer Cell 33, 676-689.e673 (2018).
Beroukhim, R. et al. The landscape of somatic copy-number alteration across human cancers. Nature 463, 899 (2010).
Zack, T. I. et al. Pan-cancer patterns of somatic copy number alteration. Nat. Genet. 45, 1134 (2013).
Akavia, U. D. et al. An integrated approach to uncover drivers of cancer. Cell 143, 1005–1017 (2010).
Ohshima, K. et al. Integrated analysis of gene expression and copy number identified potential cancer driver genes with amplification-dependent overexpression in 1,454 solid tumors. Sci. Rep. 7, 641 (2017).
Dong, G. et al. Integrative analysis of copy number and transcriptional expression profiles in esophageal cancer to identify a novel driver gene for therapy. Sci. Rep. 7, 42060–42060 (2017).
Calabrese, C. et al. Genomic basis for RNA alterations in cancer. Nature 578, 129–136 (2020).
Shao, X. et al. Copy number variation is highly correlated with differential gene expression: a pan-cancer study. BMC Med. Genet. 20, 175 (2019).
Fejzo, M. S. et al. Proteasome ubiquitin receptor PSMD4 is an amplification target in breast cancer and may predict sensitivity to PARPi. Genes Chromosom. Cancer 56, 589–597 (2017).
Wang, H. et al. PSMB4 overexpression enhances the cell growth and viability of breast cancer cells leading to a poor prognosis. Oncol. Rep. rts 40, 2343–2352 (2018).
Zhang, X. et al. Overexpression of SCAMP3 is an indicator of poor prognosis in hepatocellular carcinoma. Oncotarget 8, 109247–109257 (2017).
Orsetti, B. et al. Genetic profiling of chromosome 1 in breast cancer: mapping of regions of gains and losses and identification of candidate genes on 1q. Br. J. Cancer 95, 1439–1447 (2006).
Wang, M. & Lemos, B. Ribosomal DNA copy number amplification and loss in human cancers is linked to tumor genetic context, nucleolus activity, and proliferation. PLoS Genet. 13, e1006994 (2017).
Huang, Y. et al. Overexpressed ACBD3 has prognostic value in human breast cancer and promotes the self-renewal potential of breast cancer cells by activating the Wnt/beta-catenin signaling pathway. Exp. Cell Res. 363, 39–47 (2018).
Gupta, A. et al. PAK2-c-Myc-PKM2 axis plays an essential role in head and neck oncogenesis via regulating Warburg effect. Cell Death Dis. 9, 825–825 (2018).
Buschman, M. D., Rahajeng, J. & Field, S. J. GOLPH3 links the Golgi, DNA damage, and cancer. Cancer Res. 75, 624–627 (2015).
Xu, W. et al. Enhanced expression of cohesin loading factor NIPBL confers poor prognosis and chemotherapy resistance in non-small cell lung cancer. J. Transl. Medi. 13, 153–153 (2015).
Zhang, H., Zhang, C. F. & Chen, R. Zinc finger RNA-binding protein promotes non-small-cell carcinoma growth and tumor metastasis by targeting the Notch signaling pathway. Am. J. Cancer Res. 7, 1804–1819 (2017).
González-Reymúndez, A. & Vázquez, A. I. Multi-omic signatures identify pan-cancer classes of tumors beyond tissue of origin. Sci. Rep. 10, 8341 (2020).
Kang, J. U. Characterization of amplification patterns and target genes on the short arm of chromosome 7 in early-stage lung adenocarcinoma. Mol. Med. Rep. 8, 1373–1378 (2013).
Dalmasso, C. et al. Patterns of chromosomal copy-number alterations in intrahepatic cholangiocarcinoma. BMC Cancer 15, 126–126 (2015).
Ryan, S. D. et al. Up-regulation of the mitotic checkpoint component Mad1 causes chromosomal instability and resistance to microtubule poisons. Proc. Natl. Acad. Sci. 109, E2205–E2214 (2012).
Wang, H. et al. Translation initiation factor eIF3b expression in human cancer and its role in tumor growth and lung colonization. Clin. Cancer Res. 19, 2850–2860 (2013).
Hu, Z.-Y. et al. AKAP-9 promotes colorectal cancer development by regulating Cdc42 interacting protein 4. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 1862, 1172–1181 (2016).
Chen, J. et al. CSN6 drives carcinogenesis by positively regulating Myc stability. Nat. Commun. 5, 5384–5384 (2014).
Butler, J. S. et al. Low expression of ASH2L protein correlates with a favorable outcome in acute myeloid leukemia. Leuk. Lymphoma 58, 1207–1218 (2017).
Nikolsky, Y. et al. Genome-wide functional synergy between amplified and mutated genes in human breast cancer. Cancer Res. 68, 9532–9540 (2008).
Chen, X. N. et al. From amplification to gene in thyroid cancer: a high-resolution mapped bacterial-artificial-chromosome resource for cancer chromosome aberrations guides gene discovery after comparative genome hybridization. Am. J. Hum. Genet. 63, 625–637 (1998).
Shi, H. et al. YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 27, 315–328 (2017).
Chin, S. F. et al. High-resolution aCGH and expression profiling identifies a novel genomic subtype of ER negative breast cancer. Genome Biol. 8, R215 (2007).
Kang, J. U. et al. High frequency of genetic alterations in non-small cell lung cancer detected by multi-target fluorescence in situ hybridization. J Korean Med Sci 22 Suppl, S47–S51 (2007).
Mahmood, S. F. et al. A siRNA screen identifies RAD21, EIF3H, CHRAC1 and TANC2 as driver genes within the 8q23, 8q24.3 and 17q23 amplicons in breast cancer with effects on cell growth, survival and transformation. Carcinogenesis 35, 670–682 (2013).
Tian, Z. et al. Atypical ubiquitin-binding protein SHARPIN promotes breast cancer progression. Biomed. Pharmacother. 119, 109414 (2019).
Hokonohara, K. et al. Involvement of MAF1 homolog, negative regulator of RNA polymerase III in colorectal cancer progression. Int. J. Oncol. 54, 1001–1009 (2019).
Kasugai, Y. et al. Identification of CCND3 and BYSL as candidate targets for the 6p21 amplification in diffuse large B-cell lymphoma. Clin. Cancer Res. 11, 8265–8272 (2005).
Wu, B., Qi, R., Liu, X., Qian, L. & Wu, Z. Rab18 overexpression promotes proliferation and chemoresistance through regulation of mitochondrial function in human gastric cancer. OncoTargets Ther. 11, 7805–7820 (2018).
Kwok, S. C., Kumar, S. & Dai, G. Characterization of a variant of ERGIC2 transcript. DNA Cell Biol. 33, 73–78 (2014).
Hirasawa, A. et al. Association of 17q21-q24 gain in ovarian clear cell adenocarcinomas with poor prognosis and identification of PPM1D and APPBP2 as likely amplification targets. Clin. Cancer Res. 9, 1995–2004 (2003).
Bhatnagar, S. et al. TRIM37 is a new histone H2A ubiquitin ligase and breast cancer oncoprotein. Nature 516, 116 (2014).
Toffoli, S. et al. Identification by array comparative genomic hybridization of a new amplicon on chromosome 17q highly recurrent in BRCA1 mutated triple negative breast cancer. Breast Cancer Res. BCR 16, 466–466 (2014).
Kuuselo, R. et al. 19q13 amplification is associated with high grade and stage in pancreatic cancer. Genes Chromosom. Cancer 49, 569–575 (2010).
Kanemoto, K. et al. Genomic landscape of experimental bladder cancer in rodents and its application to human bladder cancer: gene amplification and potential overexpression of Cyp2a5/CYP2A6 are associated with the invasive phenotype. PLoS ONE 11, e0167374 (2016).
Munkley, J. et al. Androgen-dependent alternative mRNA isoform expression in prostate cancer cells. F1000Res 7, 1189–1189 (2018).
Kumaran, M. et al. Germline copy number variations are associated with breast cancer risk and prognosis. Sci. Rep. 7, 14621 (2017).
Dolezal, J. M., Dash, A. P. & Prochownik, E. V. Diagnostic and prognostic implications of ribosomal protein transcript expression patterns in human cancers. BMC Cancer 18, 275–275 (2018).
Aslan, B. et al. The ZNF304-integrin axis protects against anoikis in cancer. Nat. Commun. 6, 7351 (2015).
Correa, B. R. et al. Functional genomics analyses of RNA-binding proteins reveal the splicing regulator SNRPB as an oncogenic candidate in glioblastoma. Genome Biol. 17, 125–125 (2016).
Kim, J. J., Lee, S. B., Park, J. K. & Yoo, Y. D. TNF-α-induced ROS production triggering apoptosis is directly linked to Romo1 and Bcl-XL. Cell Death Differ. 17, 1420 (2010).
Fejzo, M. S. et al. Amplification target ADRM1: role as an oncogene and therapeutic target for ovarian cancer. Int. J. Mol. Sci. 14, 3094–3109 (2013).
Bagchi, A. & Mills, A. A. The quest for the 1p36 tumor suppressor. Cancer Res. 68, 2551–2556 (2008).
Lim, S. K. & Gopalan, G. Antizyme1 mediates AURKAIP1-dependent degradation of Aurora-A. Oncogene 26, 6593 (2007).
Ali, A. M. et al. FAAP20: a novel ubiquitin-binding FA nuclear core-complex protein required for functional integrity of the FA-BRCA DNA repair pathway. Blood 119, 3285–3294 (2012).
Sun, Y. et al. MIIP haploinsufficiency induces chromosomal instability and promotes tumour progression in colorectal cancer. J. Pathol. 241, 67–79 (2017).
Li, Y. et al. MAD2L2 inhibits colorectal cancer growth by promoting NCOA3 ubiquitination and degradation. Mol. Oncol. 12, 391–405 (2018).
Kluth, M. et al. Genomic deletion of MAP3K7 at 6q12-22 is associated with early PSA recurrence in prostate cancer and absence of TMPRSS2:ERG fusions. Mod. Pathol. 26, 975 (2013).
Willan, J. et al. ESCRT-III is necessary for the integrity of the nuclear envelope in micronuclei but is aberrant at ruptured micronuclear envelopes generating damage. Oncogenesis 8, 29 (2019).
Karnik, P., Chen, P., Paris, M., Yeger, H. & Williams, B. R. G. Loss of heterozygosity at chromosome 11p15 in Wilms tumors: identification of two independent regions. Oncogene 17, 237 (1998).
Tapia-Laliena, M. Á et al. Cullin 5 is a novel candidate tumor suppressor in renal cell carcinoma involved in the maintenance of genome stability. Oncogenesis 8, 4–4 (2019).
Mertens, D. et al. Down-regulation of candidate tumor suppressor genes within chromosome band 13q14.3 is independent of the DNA methylation pattern in B-cell chronic lymphocytic leukemia. Blood 99, 4116–4121 (2002).
Zhang, C. et al. Downregulation of ZC3H14 driven by chromosome 14q31 deletion promotes hepatocellular carcinoma progression by activating integrin signaling. Carcinogenesis 40, 474–486 (2018).
Rakha, E. A., Armour, J. A. L., Pinder, S. E., Paish, C. E. & Ellis, I. O. High-resolution analysis of 16q22.1 in breast carcinoma using DNA amplifiable probes (multiplex amplifiable probe hybridization technique) and immunohistochemistry. Int. J. Cancer 114, 720–729 (2005).
Wei, J.-X. et al. Vps4A functions as a tumor suppressor by regulating the secretion and uptake of exosomal microRNAs in human hepatoma cells. Hepatology (Baltimore, Md.) 61, 1284–1294 (2015).
Dewez, M. et al. The conserved Wobble uridine tRNA thiolase Ctu1–Ctu2 is required to maintain genome integrity. Proc. Natl. Acad. Sci. 105, 5459–5464 (2008).
Chuensumran, U., Wongkham, S., Pairojkul, C., Chauin, S. & Petmitr, S. Prognostic value of DNA alterations on chromosome 17p13.2 for intrahepatic cholangiocarcinoma. World J. Gastroenterol. 13, 2986–2991 (2007).
Byun, D.-S. et al. Hypermethylation of XIAP-associated factor 1, a putative tumor suppressor gene from the 17p13.2 locus, in human gastric adenocarcinomas. Cancer Res. 63, 7068–7075 (2003).
Buffet, A. et al. Germline mutations in the mitochondrial 2-oxoglutarate/malate carrier SLC25A11 gene confer a predisposition to metastatic paragangliomas. Cancer Res. 78, 1914–1922 (2018).
Packer, R. J. & Hoffman, E. P. Neuro-oncology: understanding the molecular complexity of medulloblastoma. Nat. Rev. Neurol. 8, 539–540 (2012).
Islam, M. M. et al. HDAC10 as a potential therapeutic target in ovarian cancer. Gynecol. Oncol. 144, 613–620 (2017).
Kruse, J.-P. & Gu, W. p53 aerobics: The major tumor suppressor fuels your workout. Cell Metab. 4, 1–3 (2006).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. 102, 15545–15550 (2005).
Jin, K. et al. Long non-coding RNA PVT1 interacts with MYC and its downstream molecules to synergistically promote tumorigenesis. Cell Mol. Life Sci. 76, 4275–4289 (2019).
Gu, Y. et al. The oncogenic potential of the centromeric border protein FAM84B of the 8q24.21 gene desert. GenesGenes (Basel) 11, 312 (2020).
Grossman, R. L. et al. Toward a shared vision for cancer genomic data. N. Engl. J. Med. 375, 1109–1112 (2016).
Daub, C. O., Steuer, R., Selbig, J. & Kloska, S. Estimating mutual information using B-spline functions—an improved similarity measure for analysing gene expression data. BMC Bioinformatics 5, 118 (2004).
Acknowledgements
The results presented here are in whole based upon data generated by the TCGA Research Network: https://cancergenome.nih.gov.
Author information
Authors and Affiliations
Contributions
G.W. and D.A. conceived and designed the experiments, performed the experiments, analyzed the data, contributed analysis tools, and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, G., Anastassiou, D. Pan-cancer driver copy number alterations identified by joint expression/CNA data analysis. Sci Rep 10, 17199 (2020). https://doi.org/10.1038/s41598-020-74276-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-74276-6
This article is cited by
-
Completing a genomic characterisation of microscopic tumour samples with copy number
BMC Bioinformatics (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.