Abstract
The Caspase activation and recruitment domain 8 (CARD8) protein is a component of innate immunity and overexpression of CARD8 mRNA was previously identified in atherosclerosis. However, very little is known about the regulation of CARD8 in endothelial cells and atherosclerosis. The aim of this study was to investigate CARD8 in the regulation of cytokine and chemokine expression in endothelial cells. Sections of human atherosclerotic lesions and non-atherosclerotic arteries were immunostained for CARD8 protein. Expression of CARD8 was correlated to mediators of inflammation in atherosclerotic lesions using Biobank of Karolinska Endarterectomies microarray data. The CARD8 mRNA was knocked-down in human umbilical vein endothelial cells (HUVECs) in vitro, followed by quantitative RT-PCR analysis and OLINK Proteomics. Endothelial and smooth muscle cells in arterial tissue expressed CARD8 and CARD8 correlated with vWF, CD163 and the expression of inflammatory genes, such as CXCL1, CXCL6 and PDGF-A in plaque. Knock-down of CARD8 in HUVECs significantly altered proteins involved in inflammatory response, such as CXCL1, CXCL6, PDGF-A, MCP-1 and IL-6. The present study suggest that CARD8 regulate the expression of cytokines and chemokines in endothelial cells and atherosclerotic lesions, suggesting that CARD8 plays a significant role in endothelial activation.
Similar content being viewed by others
Introduction
Inflammation is a key component in the pathophysiology of atherosclerosis and involves numerous complex inflammatory cascades contributing to the inflammatory milieu in the atherosclerotic lesions. The caspase recruitment domain (CARD) was initially identified as a protein–protein interaction motif in the regulation of apoptosis1 and is also known for its function as scaffolding molecule to induce inflammation by activating NF-κB2,3. During the past decade, several CARD containing proteins, such as Nod1, Nod2, CARD10, Bcl10, CARD11 and CARD14 have been identified, to regulate activation of NF-κB3 via association with different adaptor proteins; CARD6, NOD1 and NOD2 recruit RIPK2, whereas CARD9, CARD10, CARD11 and CARD14 requires recruitment of BCL10 to activate NF-κB4,5,6,7,8,9.
One of the CARD containing proteins that has previously attracted focus is CARD8 (also known as TUCAN/CARDINAL/NDDP1). The CARD8 has in earlier studies been associated with a possible role in the NLRP3 inflammasome complex and has been found over-expressed in atherosclerotic lesions10. The CARD8 gene has been extensively studied in relation to the C10X polymorphism, and several studies have suggested a possible association with various inflammatory diseases, although ambiguous results exist11. In cardiovascular disease (CVD), the minor variant of the C10X polymorphism has previously been associated with lower expression of CARD8 mRNA levels in atherosclerotic plaques and to lower levels of CRP and MCP-1 in serum, indicating that CARD8 may aggravate the atherosclerotic process by promoting inflammation10. However, the CARD8 regulatory mechanism is still not well understood. Unlike the other CARD proteins, CARD8 has been shown to inhibit NF-κB activation by interacting with IκB kinase complex (IKKγ)12. The CARD8 protein has also been suggested to inhibit the NOD2 dependent inflammatory response in epithelial cells and NLRP3 dependent IL-1β release in human monocyte derived macrophages13,14. On the other hand, earlier studies showed that CARD8 does not affect the IL-1β production and release in aortic smooth muscle cells (AoSMCs)15. Due to the fact that CARD8 is overexpressed in plaque, we hypothesize that CARD8 is important for the immune modulation in atherosclerosis. The aim of the present study is to examine the expression of CARD8 in human atherosclerotic lesion and to elucidate the role of CARD8 in the regulation of inflammatory proteins in endothelial cells and atherosclerotic lesions.
Results
CARD8 expression in healthy vessels and human carotid atherosclerotic lesions
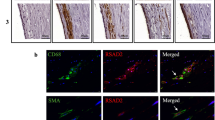
We examined CARD8 expression in non-atherosclerotic artery and carotid lesions using immunohistochemistry. In the non-atherosclerotic vessels, CARD8 expression was primarily detected in the endothelial cells (Fig. 1). Also, smooth muscle cells (SMCs) in the tunica media stained positive for CARD8. In atherosclerotic carotid lesions, CARD8 was detected in the endothelial layer, smooth muscle cells and CD68 positive macrophages (Fig. 2), suggesting that the immune cells together with vascular cells contribute to the increased expression of CARD8 in the human atherosclerotic lesion. In addition, the CARD8 expression was identified in the intimal region of atherosclerotic carotid lesions and the expression was mainly localized in smooth muscle cells and CD68 positive cells. In microarray data from the BiKE study, expression of CARD8 positively correlated with the macrophage marker CD163 (r = 0.66; p < 0.00001) and von Willebrand factor (VWF; r = 0.45; p < 0.00001) endothelial marker (Fig. 3).
Immunostaining of CARD8 in non-atherosclerotic arteries. CARD8 expression was found in the endothelial layer of intima and smooth muscle cells in the media of non-atherosclerotic artery (Left: Artery from colon tissue, Magnification, × 30; Right: Popliteal artery, Magnification, × 10). Scale bar: 100 μm. Full images are available as Supplementary material.
Representative image of immunostaining of human carotid atherosclerotic plaque. Expression of CARD8 staining (brown/DAB) was evident in endothelial cells is shown using the arrows (A), moderately in smooth muscle cells (red/SMA) (as shown with the arrow (B) and CD68 (red) positive macrophages (as shown with the arrow (C). Expression of CARD8 staining (brown/DAB) in the SMA (D) and CD68 (E) positive cells in the intimal region of lesions. Left Panel: Magnification, × 2 and Scale bar: 500 μm; Right Panel: Magnification, × 30 (D&E × 20) and Scale bar: 50 μm. Full images are available as Supplementary material.
Correlation analysis using microarray data from BiKE study. CARD8 expression positively correlated with the expression of CD163 (A), and VWF (B), in human carotid atherosclerotic plaques. Solid line represent the trend line and the dashed lines represent the 95% confidence intervals for the trend lines.
Knock down efficiency and subcellular localization of CARD8 in HUVECs
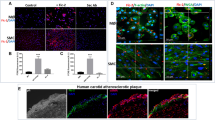
To investigate the role of CARD8 in HUVECs, the expression of CARD8 gene was silenced in HUVECs using siRNA and the efficiency of knock down was measured in terms of CARD8 mRNA and protein. The levels of CARD8 mRNA and protein expression were significantly reduced (p < 0.0001 for mRNA and protein respectively; Fig. 4). The CARD8 expression was found both in the cytoplasm and nucleus of HUVECs (Fig. 5). A substantial reduction in CARD8 expression was evident in the CARD8 siRNA treated cells compared to the control siRNA treated cells, thereby also confirming the efficiency of CARD8 knock down in the CARD8 siRNA treated cells.
Expression of CARD8 mRNA and protein in control and CARD8 knock down HUVECs. The knock down of CARD8 significantly reduced the mRNA (A) and protein levels (B) of CARD8. Representative protein bands are shown in (C). The full image of (C) is shown as Supplementary data. Data is presented as mean ± SD for n = 3–6 in each group. p value ****p < 0.0001 is compared to control.
Subcellular localization of CARD8 in HUVEC. CARD8 expression in HUVECs treated with control/scramble siRNA (A) or CARD8 siRNA (B). CARD8 expression is indicated in green, f-actin in red and nucleus are stained blue. Magnification, × 40 and Scale bar: 20 μm. Full images are available as Supplementary material.
CARD8 regulates the expression of inflammatory proteins in HUVECs
The effect of CARD8 knock-down on the expression of cytokines and chemokines was screened using three different OLINK Proseek Multiplex Assay panels; CVDII, CVDIII and Inflammation. Protein from lysate was analyzed on CVD II and CVD III panels, and protein from the culture medium was analyzed on Inflammation panel. In CVD II and CVD III panels, 46/92 and 49/92 proteins respectively, were detected in HUVECs (Supplementary Table S1). In total, 25 proteins out of 92 were detected in the cell medium using the Inflammation panel.
Knock down of CARD8 in HUVECs significantly upregulated the levels of several proteins in the lysate, including ANG1, IL17-D, BMP6, TM, t-PA, AXL, PAR1, TFF3, LDL receptor, ALCAM, PDGF-A, MMP2, SHPS1, EPHB4, CTSZ, LTBR, TNFR1, IL-6RA, ICAM2, Ep-CAM and IGFBP7, all with FDR ≤ 10% (Fig. 6A,B, Supplementary Table S2).
Differentially regulated proteins after CARD8 knock down (CARD8 KD) in HUVECs using Olink proteomics panels. Volcano plot displaying differential protein expression in the lysate (CVD II (A) and CVDIII (B) panels) and in the culture medium [Inflammation panel (C)]. Colors represent FDR levels (red, FDR ≤ 1%; green, FDR ≤ 5%; blue, FDR ≤ 10%; black, FDR > 10%). The labeled dots represent proteins that were differentially expressed in CARD8 knock down versus control HUVECs (FDR ≤ 10%). (D) The protein–protein interaction network as analyzed by String software. Proteins in the STRING software corresponds to the following: TNFRSF10B = TRAIL-R2; F2R = PAR-1; THBD = TM; TNFRSF1A = TNF-R1; TNFSF12 = TWEAK; HSPB1 = HSP27; CCL2 = MCP-1; PLAT = tPA; CXCL8 = IL-8; EIF4EBP1 = 4E-BP1; CCL7 = MCP3; ANGPT1 = ANG1: LDLR = LDL receptor; SIRPA = SHPS1; TNFRSF11B = OPG; PLAU = uPA. The red, violet and green nodes represents proteins involved in inflammatory response, cytokine-mediated signaling pathway and immune system process respectively. The colored lines represent the different possible association between the proteins. A red line indicates the presence of fusion evidence; a green line indicates neighborhood evidence; a blue line indicates co-occurrence evidence; a purple line indicates experimental evidence; a yellow line indicates text-mining evidence; a light blue line indicates database evidence; and a black line indicates co-expression evidence.
None of the proteins in the medium were found upregulated in the Inflammation panel after CARD8 knock down in HUVECs (FDR ≤ 10%; Fig. 6C). In addition to this, knock down of CARD8 in HUVECs resulted in reduction of IL-6, CXCL1, HSP27, TRAIL-R2, IL-18, PTX3, IL-27 and MCP-1 in the lysate (FDR ≤ 10%; Fig. 6A,B).
In the medium, IL-6, IL-18R1, CCL20, CXCL6, VEGF-A, OPG, MCP-1, IL-8, CD40, ADA, CXCL1, MCP3, TWEAK, 4E-BP1, MMP-1, MMP-10, CSF1, uPA, TNRFSF9 and CXCL5 were reduced after CARD8 knock down (FDR ≤ 10%; Fig. 6C).
Levels of inflammatory cytokines CXCL1, MCP-1 and IL-6 were significantly reduced both in the lysate and culture medium upon CARD8 knock down. Next, we used the STRING software to derive an interaction network of significantly altered proteins. Gene Ontology analysis identified several enriched categories including inflammatory response, cytokine-mediated signaling pathway and immune system process pathways, suggesting the important role of CARD8 in mediating inflammation in the vessel wall (Fig. 6D).
CARD8 regulates pro-inflammatory cytokines
To investigate if CARD8 knock down affects the transcription of inflammatory proteins, we examined the expression of some selective up- and downregulated proteins on the gene level. The mRNA levels of CXCL1, IL6, CXCL6, MCP-1, ALCAM, PDGFA and IL-17D were analyzed using qRT-PCR. The expression of CXCL1, IL6, CXCL6 and MCP-1 mRNAs were significantly reduced in CARD8 knock-down cells compared to control cells (CXCL1, p = 0.0034; IL6, p < 0.0001; CXCL6, p < 0.0001; MCP-1, p = 0.0066; Fig. 7). The expression of PDGFA was significantly increased in the CARD8 knock-down cells as compared to the control cells (PDGFA, p = 0.0066; Fig. 7). The expression of IL-17D and ALCAM remained unaltered in the transcriptional level (data not shown). These findings indicate that CARD8 plays a role in the regulation of these inflammatory proteins.
Gene expression levels of selected genes regulated by CARD8. The knock down of CARD8 significantly reduced the expression of CXCL1 (p = 0.0034), IL6 (p < 0.0001), CXCL6 (p < 0.0001), and MCP-1 (p = 0.0066), when compared to the control. The expression of PDGF-A (p = 0.0066) was significantly upregulated after the knock down of CARD8 when compared to control. Data are representative of samples from 3 independent experiments and displayed as mean ± SD.
CARD8 expression correlates with the mRNA expression of genes encoding inflammatory markers in the BiKE study
We further investigated the correlation of CXCL1, IL6, CXCL6, MCP-1, ALCAM, PDGF-A and IL-17D with CARD8 in human carotid plaques using microarray data from the BiKE study. Expression of CARD8 correlated significantly with CXCL1 (r = 0.48; p = 0.000000088), CXCL6 (r = 0.31; p = 0.0022), and PDGFA (r = −0.38, p = 0.000062; Fig. 8). Also, a weak correlation was found between the expression of CARD8 and MCP-1 (r = 0.28; p = 0.011), IL6 (r = 0.29; p = 0.00082), and ALCAM (r = 0.26, p = 0.026; Fig. 8). Positive correlation was found between the expression of CARD8 and CXCL1 and CARD8 and CXCL6 respectively (Fig. 8). A weak positive correlation was also found between the expression of CARD8 and MCP-1, CARD8 and IL6 and CARD8 and ALCAM respectively (Fig. 8). The expression of PDGFA showed a negative correlation to the expression of CARD8 (Fig. 8). No significant correlation was found between the expression of CARD8 and IL-17D (data not shown).
Correlation between gene expression of CARD8 and genes associated to inflammatory response and cell migration in human atherosclerotic lesions. CARD8 correlated with (A) CXCL1 (r = 0.48, p = 0.000000088); (B) CXCL6 (r = 0.31, p = 0.0022); (C) MCP-1 (r = 0.28, p = 0.011); (D) ALCAM (r = 0.26, p = 0.026); and (E) PDGFA (r = −0.38, p = 0.000062); (F) IL6 (r = 0.29; p = 0.00082).
Discussion
In this study, we show that CARD8 protein is expressed in endothelial cells and SMCs of healthy and atherosclerotic vessels and that CARD8 may be an upstream regulator of several inflammatory cytokines and chemokines in both endothelial cells and human carotid plaques.
In a pilot study, we found that CARD8 is expressed in a variety of tissues such as brain, blood vessels, spleen, muscle, epidermis layer and hair follicles of skin (data not shown). The expression of CARD8 in endothelial cells was confirmed in microarray data generated from the Oncomine 4.4 databank (www.oncomine.org; data not shown). In the present investigation, we found expression of CARD8 protein in endothelial cells and SMCs in non-atherosclerotic vessels. We also identified CARD8 protein expression in the intimal region in SMCs and CD68 positive cells. Previously we showed that CARD8 mRNA expression was elevated in atherosclerotic lesions10. The present data complements the previous study by showing that in atherosclerotic lesions, immune cells, such as CD68 positive cells, endothelial cells and SMCs are the predominant cell types expressing CARD8 protein. We further correlated the expression of CARD8 in the human atherosclerotic lesion with the endothelial and macrophage markers with microarray data from the BiKE study. Consistent with the immunostaining, the expression of CARD8 mRNA positively correlated with the mRNA expression of vWF (endothelial marker), and macrophage marker, CD163 in atherosclerotic lesions. The expression of CARD8 was localized both in the nucleus and cytoplasm of HUVECs, which was supported by a previous study where a similar subcellular localization of CARD8 was found in MCF-7 cells transfected with GFP tagged CARD8 plasmid12. Our results are consistent with previously published analysis of gene expression pattern in atherosclerotic lesions showing that CARD8 is upregulated in human atherosclerotic lesions and is significantly correlated to genes involved in inflammatory response, including CCL2/MCP1 and CD68 suggesting a role of CARD8 in mediating inflammatory markers by macrophages in the atherosclerotic lesion16.
Knowledge about the role of CARD8 in the regulation of inflammatory markers in endothelial cells and atherosclerosis is however limited. The CARD8 belongs to the bipartite CARDs, which consists of a CARD motif and one additional motif17. In contrast to other CARD containing proteins, initial studies showed that CARD8 is an inhibitor of NF-κB activation via physical interaction with the IκB kinase complex12. Previously, CARD8 was shown to regulate IL-1β release upon LPS and ATP stimulation in human-monocyte derived macrophages13. Contradictory, we have earlier shown that CARD8 does not affect the IL-1β levels in AoSMCs15. Furthermore, knock down of CARD8 did not affect NF-κB activation in HUVECs (data not shown). In order to elucidate the role of CARD8 as a regulator of inflammation, we therefore silenced the CARD8 gene to elucidate the role of CARD8 in endothelial cells. Knockdown of CARD8 in HUVECs altered several proteins involved in inflammation and chemotaxis, such as CXCL1, IL-6, CXCL6, MCP-1, and PDGF-A. This was supported by the correlation of CARD8 expression to CXCL1, PDGFA and CXCL6 and a weak positive correlation to MCP-1 and IL6 in human atherosclerotic lesions in the BiKE study, which suggests a role of CARD8 in the regulation of these proteins in human atherosclerotic plaque.
Among the several CARD8 regulated proteins identified in the present study, there are several indications that CARD8 plays a role in atherosclerosis via its impact on downstream target genes. Knockdown of the chemokine CXCL1 has in previous studies been shown to reduce atherosclerosis in atherosclerosis-prone LDLR−/− mice18. Furthermore, CXCL6 is involved in recruitment of neutrophils, but its role in CVD remains to be elucidated19. MCP-1 is a major chemotactic protein, that acts via CCR2 receptor and is induced by modified LDL and triggers adhesion of monocytes to the endothelium20. In the ApoE knock out mouse model, deletion of MCP-1 leads to the reduction in atherosclerotic lesion size21, which may indicate on an important regulatory role for CARD8 in atherosclerosis via MCP-1. This is also consistent with our previous study showing elevated mRNA expression in atherosclerotic lesions and that lower expression of the truncated CARD8 rare variant is associated with lower levels of MCP-1 in plasma from patients with myocardial infarction10. Moreover, CARD8 regulates the expression of IL-6 in the endothelial cells. The cytokine IL-6 is known to regulate the synthesis of acute phase proteins, thereby contributing to the development of auto immune and chronic inflammatory disease22. IL-6 is also shown to induce the MCP-1 via JAK/STAT3 and PI3K/AKT pathways in human vascular endothelial cells23. Several studies indicate that blocking IL-6 signaling can be protective against inflammation24. Therefore, elevated levels of CARD8 in atherosclerosis may therefore contribute to increased inflammation in the pathogenesis of atherosclerosis by upregulation of CXCL1, CXCL6, IL-6 and possibly also MCP-1, may aggravate atherosclerosis.
In the present investigation, we also found upregulation of PDGF-A after knock down of CARD8 in HUVECs and negative correlation to CARD8 mRNA expression levels in atherosclerotic lesions in the BiKE study. Elevated levels of PDGF-A have been found in atherosclerotic lesions in ApoE mice25. In addition, PDGF-A is induced by oxidized low density lipoproteins and sheer stress in vascular smooth muscle cells and HUVECs respectively26,27. However, the role of CARD8 in the regulation of PDGF-A in atherosclerosis remains to be elucidated.
Several additional proteins were found reduced as a consequence of CARD8 knock-down, such as IL-8, TRAIL-R2, PTX3, CCL20, and MCP-3 and are involved in inflammation. Moreover, proteins like IL-8, PTX3, LDL-receptor and CCL20 are known to contribute to the development of atherosclerosis28,29,30. Our study also implicates on additional roles of CARD8, since proteins of hemostasis and wound healing, such as PAR-1, TM, tPA, uPA, TFF3 and AXL were found upregulated upon silencing of CARD8, suggesting the role of CARD8 to maintain hemostasis and wound healing process. CARD8 was also found to regulate the expression of cathepsin Z (CTSZ), a cysteine protease with exopeptidase activity regulating adhesion, migration and maturation of immune cells31. Similar to CARD8, the expression of CTSZ is significantly upregulated in human atherosclerotic lesion and was previously shown to significantly correlate to inflammatory markers, including CCL2/MCP1 and CD68 suggesting the possible role of CTSZ in inflammation16.
In conclusion, the current study shows that CARD8 protein is upregulated in atherosclerotic lesions and our data suggest that CARD8 may be involved in the regulation of the expression of cytokines and chemokines, such as CXCL1, CXCL6 and PDGF-A in vascular cells and atherosclerotic plaque, but also to other proteins related to inflammation, such as MCP-1 and IL-6. However, additional studies are warranted to elucidate the more precise mechanism of regulation of these proteins. Although we lack in vivo evidence of CARD8 as a regulator of inflammatory markers, the present study suggest a possible role for CARD8 as a novel mediator of inflammation in atherosclerotic lesions.
Methods
Immunohistochemistry
Carotid artery plaques were obtained from patients undergoing carotid endarterectomy at the Division of Thoracic and Cardiovascular Surgery, Örebro University Hospital, Sweden. Non-atherosclerotic arteries from colon tissue was obtained from the Division of Pathology, Örebro University Hospital and popliteal artery was obtained from the Karolinska Institute. The samples were used for immunostaining. The use of human atherosclerotic lesions and non-atherosclerotic arteries was ethically approved by Uppsala Regional Ethical Board (Dnr 2015/532) and the Karolinska Institute ethics committee (file number 02-147 and 2009/295-31/2) and informed written consent was obtained from all individuals. The study was ethically performed according to the guidelines of the Helsinki Declaration.
Atherosclerotic and non-atherosclerotic tissues were formalin fixed and paraffin embedded at the Department of Pathology, Örebro University Hospital, Sweden. The paraffin-embedded tissues were sectioned (4 μm) and subjected to deparaffinization and rehydration using Tissue Clear (Sakura, Alphen aan den Rijn, The Netherlands) and decreasing concentrations of ethanol. Epitope retrieval was performed in Diva decloaking buffer pH 6 (Biocare Medical, Pacheco, CA, USA) and Decloaking chamber (Biocare Medical) for 10 min at 110 °C. The staining was performed using primary antibodies against CARD8 (PAB0281; Abnova Corp, Taipei City, Taiwan; 1:1000/Nordic BioSite AB, Täby, Sweden), CD68 (NCL‐L‐CD68, Novacastra, Newcastle, UK; 1:50), VWF (M0616; Dako, Glostrup, Denmark; 1:50) and smooth muscle actin (SMA; M0851; Dako; 1:500) diluted in Da Vinci Green (Biocare Medical) and incubated for 1 h at room temperature. Subsequently, the antibodies were detected using the MACH 2 double stain HRP/AP polymer detection kit followed by visualization of CARD8 with 3,3″diaminobenzidine (DAB) and of SMA, CD68 and vWF using Warp Red. The tissue sections were counterstained with Mayer’s hematoxylin for 5 min at room temperature. Finally, the slides were dehydrated in increasing concentrations of ethanol prior to mounting with Pertex mounting medium (Histolab, Gothenburg, Sweden). The digital scanner Panoramic 250 Flash III (3DHistech, Budapest, Hungary) was used to scan the slides and micrographs were obtained from the Case Viewer using autosettings (Open source version 2.0 software; 3DHistech; https://www.3dhistech.com/).
Microarray
Human atherosclerotic plaques from 126 patients undergoing endarterectomy for ischemic cerebrovascular disease were obtained from the Biobank of Karolinska Endarterectomies (BiKE), Karolinska University Hospital, Sweden. The sampling and the baseline characteristics of BiKE study have been described previously32. The study was approved by the Karolinska Institute ethics committee (file number 02-147 and 2009/295-31/2). Written consent was obtained from all participants and the study was ethically performed according to the guidelines of the Helsinki Declaration.
The human atherosclerotic plaques from the BiKE study were analyzed for gene expression via microarray using Affymetrix HG-U133 plus 2.0 Genechip arrays32. The raw data was processed using the robust microarray average algorithm and analyzed on a log2 scale, as recommended. Detailed methodology on the mRNA extraction and microarray protocol is mentioned elsewhere32. The BiKE data has been submitted to a public repository: https://www.omicsdi.org/dataset/arrayexpress-repository/E-GEOD-21545.
Cell culture
Primary human umbilical vein endothelial cells (HUVECs; Thermo Fisher Scientific, Rockford, IL, USA), from pooled donors were cultured in VascuLife basal medium supplemented with VascuLife VEGF Life factors kit (Lifeline Cell Technology, GmbH, Troisdorf, Germany) in a humidified incubator with 5% CO2 at 37 °C. The cells between passages 4 and 8 were used in the experiment.
Transfection using siRNA in HUVECs
The HUVECs (2 × 105 cells/well) were plated in six-well plates and allowed to grow overnight and transfected with siRNA followed by incubation for 48 h. The transfection mixture comprised of CARD8 stealth RNAi (Cat. no. 53621349; Invitrogen, Carlsbad, CA, USA) or stealth RNAi siRNA Negative Control Med GC Duplex (Cat. no. 462001, Invitrogen) at a final concentration of 10 nM, and Lipofectamine 2000 (Cat. no. 11668-019, Invitrogen), diluted in Opti-MEM Reduced Serum Medium (Invitrogen) according to the manufacturer’s protocol.
Subcellular localization of CARD8 in HUVECs
HUVECs were transfected with CARD8 or control siRNA by lipofection followed by incubation for 48 h. CARD8 knock down and control HUVECs were washed gently with 1 × PBS and fixed with ice-cold 4% paraformaldehyde for 40 min at room temperature. Following the incubation, the cells were washed gently with PBS and incubated with ice-cold PBS containing 0.1% Triton-× 100 for 10 min. The cells were washed again with ice-cold PBS and blocked with 1% BSA in PBS containing 0.1% Triton-× 100 for 30 min at room temperature. After gentle rinse with ice-cold PBS, the cells were stained for CARD8 using CARD8 rabbit polyclonal antibody (1:500) diluted in PBS (Abnova, Taipei, Taiwan/Nordic BioSite) for 1 h at room temperature. The cells were washed again and incubated with Alexa Fluor 488 goat anti rabbit IgG (working concentration of 2 mg/ml, Invitrogen) at room temperature for 1 h. After rinsing with ice-cold PBS, the cells were stained for F-actin using Rhodamine phallodin (5U/200 μl) in the dark for 20 min. Cells were washed twice with PBS and the nucleus was stained using 4′,6′-diamidino-2-phenylindole hydrochloride (DAPI; Sigma, Deisenhofer, Germany) in the dark for 5 min. The cover slips were removed from the wells and mounted on to slides using mounting media and viewed under fluorescence microscope Olympus BX60 fluorescence microscope (Olympus Europe, Hamburg, Germany). Images were obtained with Olympus DP71 camera (Olympus Europe).
RNA extraction and quantitative real-time PCR (qRT-PCR) analysis
Total RNA was extracted from CARD8 knock down and control HUVECs using the E.Z.N.A Total RNA Kit I (Omega Bio-Tek, Norcross, GA, USA) in accordance to the manufacturer’s instructions. The cDNA was prepared using High Capacity cDNA reverse transcription kit (Thermo Fisher Scientific), and analyzed for the mRNA expression of chemokine CXC motif ligand 1 (CXCL1; Hs00236937), chemokine CXC motif ligand 6 (CXCL6; Hs00605742), monocyte chemotactic protein 1 (MCP-1; Hs00234140), interleukin 6 (IL6; Hs00174131), platelet-derived growth factor, alpha (PDGFA; Hs00234994), activated leukocyte cell adhesion molecule (ALCAM; Hs00977641) and peptidyl-prolyl cis–trans isomerase B (PPIB; Hs00168719) and interleukin-17D (IL-17D; Hs05007146) using TaqMan universal PCR master mix, TaqMan primers and probes and 7900HT Fast Real Time PCR (Thermo Fisher Scientific) according to the manufacturer’s instructions. The data were normalized relative to PPIB as endogenous control.
OLINK Proseek Multiplex Assay
Cell lysates and culture medium from CARD8 knock down and control HUVECs from three individual experiments were analyzed utilizing Cardiovascular II (CVDII), Cardiovascular III (CVDIII) and Inflammation panels. The panels contain a broad array of 92 established protein biomarkers each for CVD and inflammation (www.olink.com), via Olink Proseek Multiplex Assay using proximity extension assay (PEA) technology33,34 (Olink Proteomics, Uppsala, Sweden). Protein quantities were log2 transformed using Olink Wizard GenEx (MultiD Analyses, Gothenburg, Sweden). Gene ontology analysis was performed using the STRING software (version 11.0; https://string-db.org/) to derive network interaction between the significantly altered proteins.
Statistical analysis
Pearson′s method was used to calculate correlation coefficients, and all correlations were adjusted for multiple comparisons using the Benjamini–Hochberg false-discovery rate (FDR) method. Experimental data are represented as mean ± SEM. Student’s T-test was used to analyze the statistical difference between two groups. A p value ≤ 0.05 was considered as the level of significance. For the statistical comparison of two groups, multiple T-test with FDR threshold of 1%, 5% and 10% (two-stage linear step-up procedure of Benjamini et al.35), with Q = 1%, Q = 5% and Q = 10%) were performed using GraphPad Prism (version 5.01; GraphPad Software, San Diego, CA, USA; https://www.graphpad.com/scientific-software/prism/).
Change history
19 April 2021
A Correction to this paper has been published: https://doi.org/10.1038/s41598-021-88467-2
References
Hofmann, K., Bucher, P. & Tschopp, J. The CARD domain: a new apoptotic signalling motif. Trends Biochem. Sci. 22, 155–156 (1997).
Blonska, M. & Lin, X. NF-kappaB signaling pathways regulated by CARMA family of scaffold proteins. Cell Res. 21, 55–70 (2011).
Jiang, C. Y. & Lin, X. Regulation of NF-kappa B by the CARD proteins. Immunol. Rev. 246, 141–153 (2012).
Dufner, A., Pownall, S. & Mak, T. W. Caspase recruitment domain protein 6 is a microtubule-interacting protein that positively modulates NF-kappa B activation. Proc. Natl. Acad. Sci. USA 103, 988–993 (2006).
Ogura, Y. et al. Nod2, a Nod1/Apaf-1 family member that is restricted to monocytes and activates NF-kappa B. J. Biol. Chem. 276, 4812–4818 (2001).
Kobayashi, K. et al. RICK/Rip2/CARDIAK mediates signalling for receptors of the innate and adaptive immune systems. Nature 416, 194–199 (2002).
Bertin, J. et al. CARD11 and CARD14 are novel caspase recruitment domain (CARD)/membrane-associated guanylate kinase (MAGUK) family members that interact with BCL10 and activate NF-kappa B. J. Biol. Chem. 276, 11877–11882 (2001).
Bertin, J. et al. CARD9 is a novel caspase recruitment domain-containing protein that interacts with BCL10/CLAP and activates NF-kappa B. J. Biol. Chem. 275, 41082–41086 (2000).
Wang, L. et al. CARD10 is a novel caspase recruitment domain/membrane-associated guanylate kinase family member that interacts with BCL10 and activates NF-kappa B. J. Biol. Chem. 276, 21405–21409 (2001).
Paramel, G. V. et al. CARD8 gene encoding a protein of innate immunity is expressed in human atherosclerosis and associated with markers of inflammation. Clin. Sci. (Lond.) 125, 401–407 (2013).
Paramel, G. V., Sirsjo, A. & Fransen, K. Role of genetic alterations in the NLRP3 and CARD8 genes in health and disease. Mediat. Inflamm. 2015, 846782 (2015).
Bouchier-Hayes, L. et al. CARDINAL, a novel caspase recruitment domain protein, is an inhibitor of multiple NF-kappa B activation pathways. J. Biol. Chem. 276, 44069–44077 (2001).
Ito, S., Hara, Y. & Kubota, T. CARD8 is a negative regulator for NLRP3 inflammasome, but mutant NLRP3 in cryopyrin-associated periodic syndromes escapes the restriction. Arthritis Res. Ther. 16, R52 (2014).
von Kampen, O. et al. Caspase recruitment domain-containing protein 8 (CARD8) negatively regulates NOD2-mediated signaling. J. Biol. Chem. 285, 19921–19926 (2010).
Tangi, T. N., Elmabsout, A. A., Bengtsson, T., Sirsjo, A. & Fransen, K. Role of NLRP3 and CARD8 in the regulation of TNF-alpha induced IL-1beta release in vascular smooth muscle cells. Int. J. Mol. Med. 30, 697–702 (2012).
Puig, O. et al. A gene expression signature that classifies human atherosclerotic plaque by relative inflammation status. Circ. Cardiovasc. Gene 4, 595-U248 (2011).
Bouchier-Hayes, L. & Martin, S. J. CARD games in apoptosis and immunity. Embo Rep. 3, 616–621 (2002).
Boisvert, W. A. et al. Up-regulated expression of the CXCR2 ligand KC/GRO-alpha in atherosclerotic lesions plays a central role in macrophage accumulation and lesion progression. Am. J. Pathol. 168, 1385–1395 (2006).
Gijsbers, K. et al. GCP-2/CXCL6 synergizes with other endothelial cell-derived chemokines in neutrophil mobilization and is associated with angiogenesis in gastrointestinal tumors. Exp. Cell Res. 303, 331–342 (2005).
Cushing, S. D. et al. Minimally modified low-density-lipoprotein induces monocyte chemotactic protein-1 in human endothelial-cells and smooth-muscle cells. Proc. Natl. Acad. Sci. USA 87, 5134–5138 (1990).
Gu, L. et al. Absence of monocyte chemoattractant protein-1 reduces atherosclerosis in low density lipoprotein receptor-deficient mice. Mol. Cell 2, 275–281 (1998).
Tanaka, T. & Kishimoto, T. Targeting Interleukin-6: all the way to treat autoimmune and inflammatory diseases. Int. J. Biol. Sci. 8, 1227–1236 (2012).
Zegeye, M. M. et al. Activation of the JAK/STAT3 and PI3K/AKT pathways are crucial for IL-6 trans-signaling-mediated pro-inflammatory response in human vascular endothelial cells. Cell Commun. Signal 16, 55 (2018).
Yao, X. et al. Targeting interleukin-6 in inflammatory autoimmune diseases and cancers. Pharmacol. Ther. 141, 125–139 (2014).
Wuttge, D. M., Sirsjo, A., Eriksson, P. & Stemme, S. Gene expression in atherosclerotic lesion of ApoE deficient mice. Mol. Med. 7, 383–392 (2001).
Zwijsen, R. M., Japenga, S. C., Heijen, A. M., van den Bos, R. C. & Koeman, J. H. Induction of platelet-derived growth factor chain A gene expression in human smooth muscle cells by oxidized low density lipoproteins. Biochem. Biophys. Res. Commun. 186, 1410–1416 (1992).
Hsieh, H. J., Li, N. Q. & Frangos, J. A. Shear stress increases endothelial platelet-derived growth factor mRNA levels. Am. J. Physiol. 260, H642-646 (1991).
Braunersreuther, V., Mach, F. & Steffens, S. The specific role of chemokines in atherosclerosis. Thromb. Haemost. 97, 714–721 (2007).
Goldstein, J. L. & Brown, M. S. The LDL receptor. Arterioscler. Thromb. Vasc. Biol. 29, 431–438 (2009).
Fornai, F. et al. The inflammatory protein Pentraxin 3 in cardiovascular disease. Immun. Ageing 13, 25 (2016).
Kos, J., Vizin, T., Fonovic, U. P. & Pislar, A. Intracellular signaling by cathepsin X: molecular mechanisms and diagnostic and therapeutic opportunities in cancer. Semin. Cancer Biol. 31, 76–83 (2015).
Folkersen, L. et al. Prediction of ischemic events on the basis of transcriptomic and genomic profiling in patients undergoing carotid endarterectomy. Mol. Med. 18, 669–675 (2012).
Assarsson, E. et al. Homogenous 96-plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS ONE 9, e95192 (2014).
Lundberg, M., Eriksson, A., Tran, B., Assarsson, E. & Fredriksson, S. Homogeneous antibody-based proximity extension assays provide sensitive and specific detection of low-abundant proteins in human blood. Nucleic Acids Res. 39, e102 (2011).
Benjamini, Y., Krieger, A. M. & Yekutieli, D. Adaptive linear step-up procedures that control the false discovery rate. Biometrika 93, 491–507 (2006).
Acknowledgements
The present study was financially supported by grants from The Knowledge Foundation (HÖG17, HÖG19, HÖG15 and Synergi 18) and The Swedish Heart and Lung Foundation. We hereby thank for their kind support.
Funding
Open Access funding provided by Örebro University.
Author information
Authors and Affiliations
Contributions
G.P.V., A.S., K.F: Conceived and designed the experiments. G.P.V., G.K., A.G.E.: Performed the experiments. G.P.V., G.K., A.G.E., L.F.: Analyzed the data. U.H, P.S.O., G.P.B., A.S., K.F.: Contributed to reagents/materials/analysis tools. G.P.V., K.F.: Wrote the manuscript. A.S., K.F.: Coordinator of the study. G.K., A.G.E., L.L., U.H., P.S.O., L.F., G.P.B., A.S.: Contributed to writing of the paper. All Authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this Article was revised: The original version of this Article contained an error in the spelling of the author Gabrielle Paulsson-Berne, which was incorrectly given as Gabrielle Paulsson Berne. In addition, the author Gabrielle Paulsson-Berne was incorrectly indexed.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Paramel, G.V., Karadimou, G., Eremo, A.G. et al. Expression of CARD8 in human atherosclerosis and its regulation of inflammatory proteins in human endothelial cells. Sci Rep 10, 19108 (2020). https://doi.org/10.1038/s41598-020-73600-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-73600-4
This article is cited by
-
Association between C10X polymorphism in the CARD8 gene and inflammatory markers in young healthy individuals in the LBA study
BMC Cardiovascular Disorders (2024)
-
Vascular smooth muscle cells in response to cholesterol crystals modulates inflammatory cytokines release and promotes neutrophil extracellular trap formation
Molecular Medicine (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.