Abstract
Down syndrome (DS) is the main genetic cause of intellectual disability due to triplication of human chromosome 21 (HSA21). Although there is no treatment for intellectual disability, environmental enrichment (EE) and the administration of green tea extracts containing epigallocatechin-3-gallate (EGCG) improve cognition in mouse models and individuals with DS. Using proteome, and phosphoproteome analysis in the hippocampi of a DS mouse model (Ts65Dn), we investigated the possible mechanisms underlying the effects of green tea extracts, EE and their combination. Our results revealed disturbances in cognitive-related (synaptic proteins, neuronal projection, neuron development, microtubule), GTPase/kinase activity and chromatin proteins. Green tea extracts, EE, and their combination restored more than 70% of the phosphoprotein deregulation in Ts65Dn, and induced possible compensatory effects. Our downstream analyses indicate that re-establishment of a proper epigenetic state and rescue of the kinome deregulation may contribute to the cognitive rescue induced by green tea extracts.
Similar content being viewed by others
Introduction
Down Syndrome (DS) is the most common genetic cause of intellectual disability and is due to the presence of three copies of chromosome 21 (HSA21). However, DS cognitive impairment is still not amenable to therapy. The most important reason is that the mechanism(s) of the cognitive impairment are still not well understood. Recent evidence has shown that green tea extracts containing epigallocatechin-3-gallate (EGCG), a green tea flavonol, improve the cognitive phenotype in trisomic Ts65Dn mice, as well as in individuals with DS1,2, providing a unique opportunity to study the molecular mechanisms underlying its beneficial effects. EGCG is known for its antioxidant properties, that could counteract the oxidative stress caused by the upregulation of CuZnSOD13 in DS. However, EGCG has many other properties that could also be beneficial in DS. It directly regulates and interacts with proteins involved in the cell membrane integrity, signal transduction, transcription factors, DNA methylation, phosphorylation, mitochondrial function and autophagy4. Of interest for DS, EGCG inhibits the Dual Specificity Tyrosine-Phosphorylation-Regulated Kinase 1A (DYRK1A), one of the most important genes in the pathogenesis of DS5,6. Previous work from our group showed that EGCG partially rescues the effects of overexpression of DYRK1A on the hippocampal proteome and phosphoproteome of TgDyrk1A mice7. However, the extent to which all these mechanisms apply to a trisomy scenario is unknown.
Interestingly, EGCG shares important similarities with environmental enrichment (EE). In Ts65Dn mice, EE improved spatial learning and memory8 , visual function9, normalized brain inhibition, enhanced hippocampal synaptic plasticity, increased branching in pyramidal cells10 and rescued postnatal neurogenesis defect11.
Moreover, we recently found that the combination of EE plus EGCG treatment is more efficient than EE or EGCG alone to ameliorate age-associated cognitive impairment of old Ts65Dn mice12 and it improves corticohippocampal-dependent learning and memory deficits in young trisomic mice. Those effect are possibly the consequences of the restoration of CA1 hippocampal dendritic spine density, and mitigation of the disturbed excitation/inhibition synaptic puncta imbalance13.
To get insight into these mechanisms we analyzed changes in protein abundances and phosphorylation in Ts65Dn mice, and their disomic counterparts in baseline conditions and upon three treatments known to improve cognition in Ts65Dn: (i) green tea extract containing EGCG, (ii) environmental enrichment (EE), and (iii) their combination.
Results
Ts65Dn hippocampal (phospho-)proteome
Using quantitative mass spectrometry-based proteomics (LC–MS/MS) on whole proteome extracts from Ts65Dn and WT mice, treated or not with EGCG containing green tea extracts (“green tea”), EE, or green tea extracts + EE (n total = 40 mice), we identified 2633 proteins, and 4705 phosphopeptides belonging to 1759 phosphorylated proteins. We fitted the data into a linear model to evaluate the changes related to the genotype, to the treatments, and their interaction, setting a threshold of adjusted p-value and fold change of p < 0.05, and |log2FC| > 0.3.
Protein dysregulated in Ts65Dn are not limited to the triplicated HSA21 genes
We found a significant increase in the abundance of 48 proteins (red dots in Fig. 1A,B), 20 of which were low or absent in WT hippocampi, and significantly decreased abundance of 67 proteins (Fig. 1A,B, Table 1).
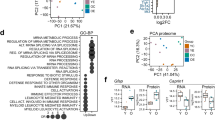
Differentially abundant proteins in each orthologous human chromosome. (a) Log2 fold change of the TS.NT − WT.NT and significance (− log10 of the adjusted p-value) of all evaluated proteins. Significant proteins are indicated in red, proteins from the trisomic regions are visualized in bigger size. The dashed horizontal red line indicates the threshold of adjusted p-value (0.05), the dashed blue vertical lines the threshold of log2FC (0.3). The significantly upregulated triplicated genes are labelled. (b) Distribution of the detected proteins along the murine chromosomes, with the y-axis showing the protein log2 fold changes. Dot colours, sizes, and labels are the same as in (a). (c) Distribution of all detected proteins with a human orthologous across the human chromosomes. (d) Distribution of the proteins whose levels are changing in trisomic mice across the human chromosomes. (e) Percentage (%) of differentially abundant proteins over the total proteins detected orthologous to each human chromosome.
Mapping all the detected protein to their respective human orthologous genes (Fig. 1C–E), revealed that their distribution across all chromosomes was as expected given the gene density of each chromosome. Since HSA21 is a gene-poor chromosome, only 19 HSA21 orthologous genes had a protein detected in our dataset. Of these, 12 are encoded by genes triplicated in Ts65Dn, of which 6 were found differentially expressed, with 5 upregulated (Table 2). When calculating the ratio of differentially abundant proteins over detected proteins, HSA21 was the one with the highest percentage (over 30%) (Fig. 1E). In Ts65Dn there are also 64 genes non-orthologous to HSA21 (mm10 release), but only the one coding for the ezrin protein (whose orthologous encoding gene is on HSA6) was detected in our proteomic analysis, and it was not differentially abundant when comparing trisomic versus wild type mice.
Kinome deregulation in Ts65Dn
We found 576 phosphopeptides—corresponding to 442 proteins—that exhibited increased abundances in Ts65Dn compared to their WT counterparts, whereas 301 phosphopeptides, mapping to 250 different proteins, were decreased in abundance (Table 3).
We aligned all the phosphopeptides with higher abundance (or exclusively present) in Ts65Dn mice creating a sequence logo of the trisomic phosphorylation motif (Supplementary Fig. S1). This showed that 502 out of 576 phosphopeptides had a phosphorylated Serine, with 176 of these also having a proline in position + 1 and 43 a proline in position − 2. The proteins changing their phosphorylation were different from those changing in abundance, with only a 10% of the dysregulated proteins changing both their levels and phosphorylation.
When analyzing the kinases with abundance or phosphorylation levels significantly changing in trisomic mice, we found 45 deregulated kinases. Of those, six kinases showed changes in abundance (2 lower and 4 higher). We also detected 12 kinases with reduced phosphorylation, which we identified mainly in the CAMK (Ca2+/calmodulin-dependent protein kinase) family (p < 0.001, Fisher Test); and 20 kinases with increased phosphorylation levels compared to the WT, belonging to the CAMK family (p < 0.001, Fisher Test), the Tyrosine Kinase-Like (TKL) (p < 0.001, Fisher Test), Ca2+/calmodulin-dependent protein kinase (CMGC) (p < 0.001, Fisher Test), protein kinase A, G, and C families (AGC) (p < 0.001, Fisher Test), Cell Kinase 1 (CK1) (p < 0.05, Fisher Test), and "Sterile" serine/threonine kinases (STE) (p < 0.05, Fisher Test) groups. Finally 7 kinases belonging to the CAMK group (p < 0.001, Fisher Test) and the AGC group (p < 0.01, Fisher Test) presented several phosphorylation sites up- or down-phosphorylated (Table 4).
The (phospho-)proteomic alterations in trisomic hippocampus are enriched in neuronal and chromatin-related categories
Having identified consistent and reproducible changes in the hippocampal proteome of Ts65Dn, we next interrogated functional associations among differentially expressed proteins. To this aim, we constructed a protein–protein interaction (PPI) network (the Ts65Dn’s network) starting from the 694 proteins that showed affected abundance or phosphorylation levels in trisomic mice, and we detected interactions among 381 proteins (seed proteins, represented in Fig. 2 as those in bigger size). To this PPI network, we also added the direct interactors ending up with 845 interacting proteins in the extended PPI network (Fig. 2). Both the seed proteins and their direct interactors were interacting more than expected by chance, suggesting that the significant changes in abundance observed in our dataset are functionally related (590 observed, versus 188 expected interactions, for seed proteins; and 3708 observed compared to 1023 expected, for the extended network). Specifically, we found a total of 1879 interactions between proteins changing their phosphorylation state; 325 interactions between proteins only changing their abundance and 207 interactions between a protein changing abundance and a protein changing phosphorylation levels.
The Ts65Dn’s protein-protein interaction network. In the network the nodes correspond to proteins changing in Ts65Dn mice (seeds) and their direct interactors, and edges correspond to known protein–protein interactions. Seed proteins are represented as bigger nodes. Circles correspond to proteins changing their abundances, squares to proteins changing their phosphorylation levels, and triangles to proteins changing both their abundance and their phosphorylation levels. The network shows nodes affected by the green tea extract (green), by EE (yellow), or by their combination (purple).
The number of interactions per protein (node degree) in the network followed a log-normal right-skewed distribution. Using this distribution, we detected 22 “hubs” in the network, defined as those appearing in the 5% right tail of the distribution, having at least 42 interactions (Supplementary Fig. S2A,B). Of these hubs, seven were seed proteins: RPS27A, SOS1, CREBBP, APP, CTNNB1, DLG4, and PLCG1 (Table 5). Of those, RPS27A, SOS1, CTNNB1, and PLCG1 had many common interactors in the network, whereas APP, CREBBP, and DLG4 had a specific subset of interactors (Supplementary Fig. S2C).
The Gene ontology categories found enriched among the deregulated proteins between wild type and trisomic mice belonged to 5 main groups: chromatin-related, neuronal projection/neuron development, microtubule/cytoskeleton, synapsis/cognition, and GTPase/Kinase activity (Supplementary Fig. S3, Table S22).
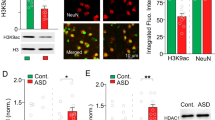
In an independent cohort of mice we confirmed both the upregulation of the neuronal protein APP, as an important hub protein in the network (Supplementary Fig. S4A,B, 2 × in Ts65Dn mice, p < 0.05) and the deregulation of chromatin-related proteins as shown by the global histone hypo-acetylation in trisomic mice (Supplementary Fig. S4C,D, p < 0.05).
Chromatin and neuronal related categories are rescued by pro-cognitive treatments in the trisomic phosphoproteome
Our results show that the treatments (EGCG containing green tea extracts, EE and their combination) modulate common targets, many of which are altered in trisomic mice (Fig. 3). In fact, changes in the proteome and the phosphoproteome exhibit a significant overlap in all the different conditions tested (around 20–30% for the proteome and 50% for the phospho-proteome; p-value Fisher’s test < 0.05) (Fig. 3A,B).
Overlap of the differentially abundant and differentially phosphorylated proteins in trisomic mice after different treatments. (a) Heatmap showing the overlap between differentially abundant proteins across the different treatments in transgenic mice. (b) Overlap between differentially abundant phosphopeptides across the different treatments. (c) Overlap between differentially abundant proteins and proteins with differentially abundant phosphopeptides. The color-code goes from very low p-values (red) to high p-values (yellow) of the exact Fisher test. TS, ts65Dn mice. WT, wild type mice. NT, not treated. Greentea, “EGCG-containing green tea extract”. EE, Environmental Enrichment. The Szymkiewicz-Simpson overlap coefficient is printed in cyan.
Given that the changes observed in protein abundances showed minimal overlap with proteins changing their phosphorylation, the trisomy and pro-cognitive treatments affected different proteins in the proteome (abundances) and the phospho-proteome (phosphorylation levels) (Fig. 3C).
We used this Ts65Dn’s network to identify potential mechanisms of the pro-cognitive treatments by analyzing their capability to restore the deregulated nodes, both at the proteome and phosphoproteome level. We considered as “rescued”, those proteins altered in Ts65Dn mice compared to WT with abundances and/or phosphorylation levels restored partially or completely, i.e. 50–150% of the WT untreated values by at least one of the treatments (Tables 1, 2, Fig. 3 and Supplementary Fig. S5, for more details see “Methods”7).
At the proteome level, we detected 57 proteins rescued by green tea and/or EE, of which 16 proteins were common to both treatments (Fig. 4A). More than half of these 57 proteins were also rescued by the combined treatment, whereas 12 proteins were found to respond only to the green tea + EE treatment. Similarly, when analyzing the rescued phosphopeptides (Fig. 4B), 570 phosphopeptides (corresponding to 437 proteins) were rescued by either green tea and/or EE, of which 208 (142 proteins) were specific to green tea, and 99 (66 proteins) to EE. The combined treatment rescued 53% of these phosphopeptides, and further rescued another 45 phosphopeptides (belonging to 24 proteins). The list of the “rescued proteins” can be found in Supplementary Table 21.
(Phospho-)proteomic alterations in trisomic mice are partially restored by the treatments. Venn Diagrams showing (a) the overlap of proteins that exhibit restored (rescued) abundances after the treatments, and (b) the overlap of phosphopeptides that exhibit restored (rescued) abundances after the treatments (in brackets the number of phosphoproteins). (c–h) Barplot showing the number of rescued proteins (c) or phosphopeptides (d), overcorrected proteins (e) or phosphopeptides (f), and impaired proteins (g) or phosphopeptides (h), after each of the tested treatments. TS, ts65Dn mice. WT, wild type mice. NT, not treated. greentea, “EGCG-containing green tea extract”. EE, Environmental Enrichment.
Altogether, these results show that green tea and EE had a number of additive and possibly synergistic effects, as their combination (green tea + EE) was different from the individual treatments. Green tea was the treatment rescuing more phosphosites, while green tea + EE was more effective at rescuing protein abundances (Fig. 4C,D). Interestingly only a small fraction of proteins was “overcorrected” or showed an even bigger difference after treatment when compared to “physiological” WT levels (Fig. 4E–H).
We then performed a gene ontology enrichment analysis of the proteins altered by the trisomy and rescued by the treatments, to better understand the molecular processes involved. When analyzing the rescued phosphopeptides, we detected significant enrichments in all the categories found deregulated at the level of the phospho-proteome (Supplementary Fig. S6, Supplementary Table 22).
We then analyzed how the rescued proteins were distributed along the Ts65Dn network (Fig. 2). Around 75% of the seed proteins were rescued by at least one of the treatments, while only 11% was overcorrected (more than 1.5-fold compared to WT untreated levels), not sufficiently rescued (less than 0.5 fold change), or further impaired. Interestingly, five out of the seven seed proteins that are hubs of the network–RPS27A , SOS1, CTNNB1, DLG4, and PLCG1–were rescued by at least one of the treatments, suggesting that the treatments are acting on key points in the Ts65Dn’s network (Table 5).
Green tea, EE, and green tea + EE rescue the trisomic kinome
Interestingly, 3 of the 6 kinases that changed their abundance (1 downregulated, and 2 upregulated), were rescued by at least one of the treatments, including the rescue of the upregulation of Erk1. Most of the de-phosphorylated kinases (34/39) recovered their phosphorylation levels by at least one of the treatments. Overall, out of the 45 deregulated kinases, 11 were rescued by each of the three treatments, and 20 by at least two of the treatments. Green tea treatment was particularly efficient in rescuing kinases from the CAMK and TKL group, while most of the kinases of the AGC and CMGC group were rescued upon EE.
Green tea, EE, and green tea + EE effects beyond the DS network
The proteins modified by the treatments included not only those altered in trisomic mice (40% of the overall proteins responding to treatments), but also 830 proteins that were not altered when comparing trisomic versus wild type mice. This indicated that the treatments were not simply restoring the portion of the proteome altered due to the trisomy, but had a much wider effect, affecting a total of 1397 proteins. In order to delineate the overall effect of the treatments we re-analyzed all the proteins responding to the treatments, and not only the rescued portion of the proteome. The gene ontology enrichment analysis detected categories related with “synapse”/neurodevelopment, chromatin/immune systems, microtubule/cytoskeleton, and GTPase activity similar to what we found for the subset of the rescued proteins (see above). Interestingly, these enrichments were lost when removing the rescued proteins. In fact, the non-rescued proteins were not enriched in any specific function when examined alone (Supplementary Fig. S6, Supplementary Table 23).
The effects of treatment with green tea, EE, and their combination are genotype specific
We assessed if there was a genotype-specific (phospho-)proteome response to the different treatments, ie. Green tea, EE, and their combination. As detailed above, in trisomic mice the treatments were rescuing most of the nodes (Fig. 2). In WT over 74% of the nodes were modified by one of the treatments (Supplementary Fig. S7, dark green nodes) shifting the abundance and/or phosphorylation of these proteins in the Ts65Dn’s network towards levels similar to the ones observed in trisomic mice. As in trisomic mice, also in WT the treatments were affecting far more proteins than those altered by the trisomy, reaching a total of 1192 proteins changing their abundance and/or phosphorylation. GO-term analysis of these proteins showed once again similar enrichments than the ones seen in the trisomic hippocampus (Supplementary Fig. S6, Supplementary Table 23). Noteworthy, the fraction of these proteins with abundance levels shifted towards trisomic levels (dark green nodes in Supplementary Fig. 5) was enriched in synaptic pathways (Supplementary Fig. S6, Supplementary Table 23).
The effects of green tea extract, EE, and the combined treatment are genotype specific. Venn Diagram showing (a) the overlap between proteins changing their abundance or their phosphopeptide levels upon any of the treatments in TS and wild type mice; and (b) the overlap between the (phospho-)proteins having the same behavior upon one of the treatments in TS mice and WT mice, and (phospho-)proteins exhibiting a genotype-specific response to treatments. (c) Plot comparing phosphopeptide (top panel) and protein (bottom panel) fold changes upon each of the three treatments in TS mice (x-axis), and wild type mice (y-axis). Protein or phosphopeptides with a significant interaction in the contrast (TS.T − TS.NT) − (WT.T − WT.NT), where T stands for one of the treatments, and NT for “not treated”, are indicated as red dots. TS, ts65Dn mice. WT, wild type mice. greentea, “EGCG-containing green tea extract”. EE, Environmental Enrichment.
Of the 1192 proteins responding to treatments in WT mice, 818 were common to the ones responding in trisomic mice (Fig. 5A, > 68% overlap, p < 0.001, Fisher Exact Test), and were enriched in the same GO-term categories (Supplementary S6, Supplementary Table 23). To understand if the treatment had genotype-specific effects, we investigated the proteins and phosphorylation sites changing in abundance upon the different treatments in WT compared to Ts65Dn mice (TS). Specifically, we compared the abundance changes observed in Ts65Dn mice upon treatment (Ts65Dn treated-Ts65Dn untreated, or TS.T–TS.U) with the abundance changes observed in WT mice upon treatment (WT treated-WT untreated, or WT.T–WT.U). Overall, we found that 998 proteins were responding significantly different to the treatments in Ts65Dn compared to WT mice. These (phospho-)proteins were enriched in categories including “synapse-related” and other neuronal components, as well as in categories associated with learning and memory (Supplementary Fig. S6, Supplementary Table 23). Of these 998 proteins, 581 were responding to the treatments both in WT and Ts65Dn mice (58% of the 998; Fig. 5B). Interestingly, when comparing the fold changes for each treatment in WT versus trisomic mice, we observed that most proteins and phosphopeptides differentially regulated by the treatments in trisomic mice, showed opposite changes in WT mice (red dots in the top-left and bottom-right portion of the plots, Fig. 5C). This observation suggests that the treatments have an opposite effect on their levels in the two genotypes.
Part of the Ts65Dn’s network overlaps DYRK1A’s
In a previously reported proteomic analysis in mice overexpressing Dyrk1A we found that the pro-cognitive treatments were rescuing the Dyrk1A transgenic hippocampal protein network of approximately 70%7. We therefore compared the list of proteins rescued by green tea, EE, or both treatments in trisomic mice with those rescued in Dyrk1A transgenic mice. We found significant overlaps (all p-values < 0.01, Fisher exact test) with 52 proteins whose phosphorylation state was rescued by green tea, EE or their combination in both Ts65Dn and Dyrk1a overexpressing mice, though in different phospho-sites (Supplementary Fig. S8). The observation agrees with the fact that 26 seed proteins of the Ts65Dn disease network are known DYRK1A interactors (including the hub APP, SYNJ1, MAP1B, BRAF, GRIN2A, AMPH, STXBP1, and SF3B1) and 17 of them (including MAP1B, STXBP1 and BRAF) were rescued by at least of one the pro-cognitive treatments.
Discussion
Environmental enrichment (EE) and the administration of green tea extracts containing epigallocatechin-3-gallate (EGCG) improve cognition in mouse models and individuals with Down syndrome, but the underlying mechanisms are unknown. We here shed light onto the possible mechanisms underlying the effects of green tea extracts, EE and their combination on the hippocampal proteome and phosphoproteome in a Down syndrome mouse model (Ts65Dn). We detected an alteration of both protein abundances and phosphorylation levels in the hippocampus of Ts65Dn mice that was not limited to triplicated genes but extended genome-wide affecting chromatin and neuronal-related categories. Interestingly, the treatment of Ts65Dn mice with green tea, EE, and their combination was able to rescue the Ts65Dn’s network.
The (phospho-)proteome deregulation is not limited to chromosome 21
The Ts65Dn mouse model is considered one of the best models for DS because of its face validity (it recapitulates most of the cognitive and neural phenotypes detected in DS) and its wide use in previous reference studies14. It bears a translocation between chromosome 17 and 16 resulting in trisomy 16 and the triplication of approximately 2/3 of the genes orthologous to HSA21. As such, we would expect a prevalence of upregulated protein coded by the triplicated regions. However, while 48 proteins increased their abundance, even more proteins, 67, decreased their level. We detected 19 proteins whose orthologous map on HSA21 (HSA21 proteins). Of those, five were upregulated: APP, SOD1, CBR1, CBR3, and PCP4. This corresponds to 30% of the detected HSA21 proteins, a percentage much higher than other chromosomes that only reached maximum 10% of differentially abundant proteins (Fig. 1C–E). A concern related to the Ts65Dn model is that it bears some triplicated genes not orthologous to HSA21. However, out of these genes only the HSA6 protein ezrin was detected in our dataset, and this protein was not differentially abundant or phosphorylated (Table 2).
Kinome and epigenomic deregulation in Ts65Dn mice
Regarding the phosphoproteome, we detected more changes in phosphopeptide levels than in protein abundances. . Interestingly, when analyzing the proteins to which these phosphopeptide belonged, we found many in common with previous findings15, such as DLGAP4, ARVCF, GRIN2A, PCLO, DLG1, DYNC1LI1, SPI1L1, RTN4, ADD2, and SYNJ, involved in synaptic processes (transport, signalling, assembly), microtubule, and glutamate receptors.
These changes in phosphorylation levels are possibly associated with the detected differences in abundance and phosphorylation state in 44 de-regulated kinases (6 with alteration in abundance, and 38 with altered phosphorylation). These proteins belonged to different kinase families but we detected the consensus motif PXSP as the most common phosphorylation site. This is a sequence recognized by mitogen-activated protein (MAP) kinases, which is important for learning and memory16.
EE, green tea extracts and their combination were able to rescue half of the kinases with altered abundances, and over 87% of the ones with altered phosphorylation, with green tea rescuing mainly CAMK and TKL kinases, and EE, AGC and CMGC kinases.
To better understand the concerted effects of proteins changing their abundance and the ones changing their phosphorylation levels in trisomic mice we analyzed them together in a PPI Ts65Dn’s network. The proteins in the network were functionally related, as suggested by their number of interactions, higher than expected by chance. When analyzing the main hubs of the Ts65Dn network we found many proteins related to Down syndrome. The protein with most interactions was ubiquitin (RPS27A), and the disturbance of the polyubiquitination machinery is a key feature of Down syndrome neurodegeneration with accumulation of polyubiquitinated toxic proteins17. Another interesting protein is SOS1, that was less phosphorylated in Ts65Dn mice, and was rescued by EE treatment. It promotes Ras activation in the MAPK/ERK pathway and is involved in axon guidance. Interestingly, mutation in this gene leads to a developmental syndrome called Noonan syndrome18 which shares hallmarks with Down syndrome like mildly unusual facial characteristics, short stature, and heart defects. Also, CREB binding protein (CBP) phosphorylation was found only in trisomic samples and it was rescued by the treatments. This protein is an histone acetyl transferase that facilitates the transcription of CREB dependent genes, such as the immediate early genes activated during cognitive processes. Mutations in the CREBBP gene cause Rubinstein-Taybi syndrome, a condition19 that, similar to Down syndrome, is characterized by short stature, moderate to severe intellectual disability, and distinctive facial features. As expected, APP abundance was increased in trisomic samples, and it was a hub in the Ts65Dn network. APP is one of the most important Down syndrome candidate gene, whose triplication is responsible for neurodegeneration and early onset Alzheimer disease in individuals with Down syndrome. Another hub of the network, Catenin Beta 1, was found phosphorylated only in trisomic mice and rescued upon our treatments. Interestingly the Wnt/β-catenin pathway is downregulated in the hippocampus of adult DS individuals with Alzheimer’s disease20. DLG4, a scaffolding protein present in the post-synaptic density implicated in glutamatergic synaptic functions21, was found differentially phosphorylated in Ts65Dn and rescued by our treatments. Finally, Phospholipase C, gamma 1 phosphorylation was not detected in trisomic samples but was present after treatment. APP, CREBBP, DLG4 formed independent modules in the network, while SOS1, beta-catenin, ubiquitin and PLCG1 formed a fourth module, indicating that they were implicated in common molecular mechanisms.
Treatment effects on the hippocampal proteome and phosphoproteome
Since green tea-extracts and EE improve cognition in Ts65Dn mice1,2 we wanted to elucidate the molecular mechanisms responsible for these improvements.
In our (phospho-)proteomic profiles, green tea extracts and EE not only rescued five out of the seven seed protein hubs but also more than 70% of the seed proteins. We detected five main categories that were compromised in trisomic mice and rescued by our pro-cognitive treatments: chromatin-related, neuronal projection/neuron development, microtuble/cytoskeleton, synapsis/cognition, and GTPase/Kinase activity.
Interestingly, green tea extracts rescued mainly protein phosphorylation, while the combination of green tea extracts and EE acted more on protein abundance. Previous studies had shown that both green tea extract with EGCG and environmental enrichment inhibit the tyrosine kinase DYRK1A5, which regulates the phosphorylation of many important proteins in the brain2,22. In a previous study7, we showed that both treatments could similarly rescue 70% of the Dyrk1A transgenic hippocampal (phospho-)protein network. Interestingly, 26 DYRK1A targets were seed proteins of the Ts65Dn network and more than 65% of these proteins were rescued by one of the pro-cognitive treatments. In both Dyrk1A transgenic mice and Ts65Dn mice the phosphorylation levels of 52 proteins were rescued by green tea, EE or their combination (Fig. S8).
An interesting mechanism revealed by our phosphoproteomic study was a re-balancing of the epigenetic state, which is compromised in Down syndrome23. Strikingly we observed a reduction in the levels of global histone H3 acetylation in the hippocampus of Ts65Dn mice, suggesting a more repressed chromatin that might contribute to a reduced expression of memory promoting genes , as previously proposed24.
Regarding our treatments, EE is known to trigger activity dependent transcription with activation of the transcription factor CREB and increase of global acetylation that favours cognition25. Moreover, EGCG shows a histone deacetylase inhibitor26,27 and DNA methyl transferase activity28. This hypothesis is corroborated by fact that among the impaired and rescued categories we found many chromatin-related molecular categories.
Another important question was the possible interaction between treatments, as previous behavioral studies have shown that the effect of the combination of green tea and environmental enrichment is more potent than each treatment alone12,13. Our (phospho-)protemic experiments revealed that the effects of the combination were different than the ones of the single treatments, indicating a strong interaction. Moreover, their effects extended far beyond the (phospho-) proteins impaired by the trisomy, although these “out of the network” proteins were enriched in similar categories. This indicates that green tea, EE and their combination not only rescue the deregulated proteins but also related proteins beyond the Ts65Dn network with a possible positive compensatory mechanism. We speculate that this would contribute to the improvements in cortico hippocampal-dependent learning and memory in trisomic mice once treated with green tea and environmental enrichment13. Unfortunately, the present experiments do not allow to directly correlate our proteomic and phospho-proteomic findings with cognitive levels in the same mice. However our previous study showed that, the combined treatment with EE and Green tea extracts improved corticohippocampal-dependent learning in both young and middle age Ts65Dn mice12,13. Conversely in DYRK1A transgenic mice the combined treatment did not show better results compared to EGCG alone7.
Our pro-cognitive treatment had a different effect on wild type mice than in Ts65Dn mice, confirming our previous observations in DYRK1A transgenic mice7. Around 44% of the proteins changing upon pro-cognitive treatments were common in wild type and trisomic mice, but with an opposite direction of change (Fig. 5). As such, over 70% of the nodes modified by the treatment(s) in WT hippocampi shifted their abundance and phosphorylation towards levels similar to the ones observed in trisomic mice. These changes could be interpreted as deleterious, and indicate that further studies are needed to optimize the treatment efficacy of green tea and EE.
Conclusion
The pro-cognitive potential of green tea extracts and environmental enrichment was already shown in previous works1,2,5,7,12,13, but the molecular effect on the proteome and phosphoproteome remained elusive. We here show that these pro-cognitive treatments are able to restore more than 70% of the protein deregulation in trisomic mice, and induce compensatory mechanisms by acting on proteins of the same categories. Our downstream analyses indicate as possible mechanisms both the re-establishment of a proper epigenetic state and the rescue of the kinome deregulation, possibly initiated by triplication of key genes such as DYRK1A. In light of these results, promising combinatorial therapies will boost or prolong current cognitive-enhancement for the treatment of intellectual disabilities.
Methods
Animal models
We used female mice from wild-type (WT) and Ts(1716)65Dn (Ts65Dn) mice, obtained through crossings of a B6EiC3Sn a/A-Ts (1716)65Dn (Ts65Dn) female to B6C3F1/J males (from The Jackson Laboratory, Bar Harbor, USA). Mice were bred in the Animal Facilitity of the Barcelona Biomedical Research Park (PRBB, Barcelona, Spain, in standard cages (20 × 12 × 12 cm Plexiglas). Animals were kept in small groups (2–3 animals) in a 12-h light–dark cycle (8:00–22:00 h), with constant humidity (60%) and temperature (22 ± 1 °C) with ad libitum access to food and water. Procedures were approved by the local ethical committee (Comité Ético de Experimentación Animal del PRBB (CEEA-PRBB); MDS-16-0035PR1-P2), meeting the guidelines of the local (law 32/2007) and European regulations (EU directive e no. 86/609, EU decree 2001-486) and the Standards for Use of Laboratory Animals no. A5388-01 (NIH). The CRG received autorization to work with genetically modified organisms (A/ES/05/I-13 and A/ES/05/14).
Experimental design
We used 2 months trisomic (TS) and wild type (WT) mice. Animal were whether left non-treated (NT) or treated with three different protocols:
-
Administered with green tea extract (greentea).
-
Reared under enriched environment (EE) conditions.
-
Both administered with green tea extract and reared under EE (greentea + EE).
We used five mice per group, randomly selected from each cage (40 total mice).
Pro-cognitive treatments
In order to prepare green tea extract (Mega Green Tea Extract, Decaffeinated, Life Extension, USA; EGCG content of 326.25 mg per capsule), we prepared the solution every 2–3 days in water at 0.33 mg/ml corresponding. This correspond to an average dose of 42 mg/Kg per day for one month.
For EE we used Plexiglas cage (55 × 80 × 50 cm) with toys, and platforms of different shapes, sizes, colors and textures, changing their arrangement every 3 days to maintain the novelty conditions. To increase social interactions we house 6–8 mice per cage, instead of 2–3, rearing them just after the weaning period, for one month, and before sexual maturity, to reduce territorial aggressiveness29, the animals were reared in the same cage just after the weaning period, before they reached sexual maturity.
Proteomics sample preparation
Dissected hippocampi were flash-frozen at -80 °C and then processed at the same time. We used RIPA-modified buffer for the homogeneization (50 mM tris–HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.1% sodium deoxycholate 5 mM b-glycerophosphate, 10 mM sodium fluoride, 10 mM sodium orthovanadate, and protease inhibitors from the Complete Protease Inhibitor Cocktail Roche).
Samples were sonicated with Bioruptor (Diagenode) for 5 min with 30 on/off cycles, on ice, and centrifuged for 10 min 10,000 rpm at 4 °C. The supernatants, containing proteins, were precipitated overnight at − 20 °C by adding six volumes of ice-cold acetone. We solubilized the acetone-precipitated proteins in a denaturation buffer (6 M urea and 200 mM ammonium bicarbonate in water. BCA-quantified proteins (Pierce), were reduced with dithiothreitol (DTT, 10 mM, 37 °C, 60 min), and alkylated with iodoacetamide (IAM, 20 mM, 25 °C, 30 min). We diluted samples with 200 mM ammonium bicarbonate up to 2 M urea, digested overnight with Lys-C at 37 °C, and then diluted two times and digested overnight with trypsin at 37 °C. Peptides were desalted using a C18 MicroSpin 300A silica column (The Nest Group Inc), dried using a speedvac, and dissolved in 0.1% formic acid in water.
Titanium dioxide (TiO2) phosphopeptide enrichment
Titansphere chromatography was used to enrich phosphopeptides as described previously7. Tryptic peptides were desalted, completely dried, and dissolved with 100 μl of Loading Buffer [80% ACN (vol/vol) and 6% TFA (vol/vol)] at ~ 1 μg/μl. Samples went through a constricted TiO2 loaded spin tip, previously equilibrated with Loading Buffer. To achieve a complete binding we applied 2 × 50 μl and used a centrifuge at ~ 50 g. We washed the TiO2 spin tip with 50 μl of Loading Buffer and with 50 μl of Washing Buffer [50% ACN (vol/vol) and 0.1% TFA (vol/vol)]. Finally, we eluted teh phosphopeptides from the TiO2 spin tip with 30 μl of Elution Buffer (85% NH3-H2O, pH 11.0) into a tube containing one volume of 20% formic acid. We performed a second elution in the same tube with 3 μl Elution Buffer 2 [(80% ACN (vol/vol) and 2% formic acid (vol/vol)]. The eluted phosphopeptides were dried and dissolved with 0.1% formic acid in water for being analyzed by mass spectrometry (MS).
Liquid chromatography-tandem mass spectrometry
2 μg of tryptic peptides from digested hippocampal tissue and phospho-enriched peptides from 100 μg of the same tissue were injected in an LTQ-Orbitrap Velos Pro mass spectrometer (Thermo Fisher Scientific) coupled to a nano-LC (EASY-nLC, Proxeon), for each samples. The orbitrap system’s performance was previously assessed showing coefficients of variations below 20% in single-shot technical replicates (from HeLa cells) for both proteome and phosphoproteome quantification30. Nano-LC was equipped with a reversed-phase chromatography column (of 25 cm) with an inner diameter of 75 μm, with 3 μm C18 particles (Nikkyo Technos, NTCC-360/75–3-25L), and a Nano Trap Column Acclaim PepMap100 100 μm × 2 cm C18, 5 μm, 100A (Thermo, 164199). The chromatographic gradients started at 93% of buffer A and 7% of buffer B with a flow rate of 250 nL/min during 5 min and linearly changed to 65% buffer A and 35% buffer B after 4 h. The column was washed for 16 min with 90% buffer A and 10% buffer B (Buffer A: 0.1% formic acid in water. Buffer B: 0.1% formic acid in acetonitrile) after each analysis.
The mass spectrometer was used in positive ionization mode with the nanospray voltage set at 2.2 kV and source temperature of 250 °C. We used the Ultramark 1621 for the FT mass analyzer for external calibration prior to the analyses. We used the background polysiloxane ion signal at m/z 445.1200 as lock mass. We operated the instrument in data-dependent acquisition (DDA) mode with 1 microscan at resolution of 60,000 at 400 m/z. Survey scans were recorded over a mass range of m/z 350−2000 with detection in the Orbitrap mass analyzer. We sat the auto gain control (AGC) to 106, dynamic exclusion to 60 s, and we activated the charge-state filter disqualifying singly charged peptides for fragmentation. Following each survey scan, We selected for fragmentation the top 20 most intense ions with multiple charged ions above a threshold ion count of 5,000 at normalized collision energy of 35%. We acquired in the linear ion trap Fragment ion spectra produced via collision-induced dissociation (CID) and collision-induced dissociation MultiStage activation (CID MSA) for proteome and phosphoproteome, respectively. AGC was set to 5 × 104 and isolation window of 2.0 m/z, activation time of 0.1 ms and maximum injection time of 100 ms.
Mass spectrometry data analysis
We acquired and processed the mass spectra were processed with the MaxQuant computational platform version 1.5.2.831 (RRID:SCR_014485). The MS2 spectra were searched with the Andromeda search engine32 against the Uniprot sequence database for Mus musculus (17,263 forward entries; version from July 2015). The search included cysteine carbamidomethylation as a fixed modification, and N-terminal protein acetylation and methionine oxidation as variable modifications. For the phosphoproteome analysis, we also added the phosphorylation on serine, threonine and tyrosine as variable modifications. We allowed a maximum of two mis-cleavages, 4.5 ppm as mass tolerance for precursor ions, and 0.5 Da as mass tolerance for fragment ions. The fraction of false discovery rate was set to 0.01 at both the peptide and protein level, and protein identification required at least one unique or razor peptide per each protein group.
We used MaxQuant algorithm to retrieve accurate extracted ion currents (XICs) per each peptide feature for quantification purposes. We calculated for each peptide the areas under the curve. For the statistical analysis we used the R package MSstats version 2.6.033 (RRID:SCR_014353). In some of the experimental replicates we excluded from the anaysis one of the five biological replicates, when the number of peptides identified was substantially lower than the average of the whole experiment (e.g. < 1000 peptides). Specifically: one replicate was excluded from the TS.greentea + EE and WT.EE groups in the proteome analysis, and another replicate was excluded from the TS.greentea and WT.greentea + EE groups in the phosphoproteome analysis. To ensure high confidence in our quantitative data, we only used peptides observed at least in three of the five biological replicates (or at least in two when we remained only with four biological replicates), and no imputation of missing values was performed.
Differentially expressed proteins and phosphopeptides
These analyses were peformed similarly to what we already described previously7. We performed the differential expression analysis with the MSstats package that is based on a family of linear mixed-effects models33. All downstream bioinformatics analyses were performed on proteins and phosphopeptides showing a significant change in abundance (Benjamini adjusted p-value lower than 0.05 and a log2(Fold Change) (log2FC) > 0.3 or < − 0.3). For the phosphoproteomic analysis, we considered only phosphorylation sites with a localization probability of 0.5 to avoid uncertain sites. We added to the lists of differentially abundant proteins and phosphopeptides the proteins or phosphopeptides uniquely present in one condition of the ones compared. A peptide was defined as “absent/low abundant” when it was detected in less than 0, 1 or 2 out of 5 biological replicates for a given condition (or less than 2 out of 4 biological replicates). Proteins belonging to the GO-term cell component “blood microparticle” were filtered out from all datasets before proceeding with downstream analyses, since blood contamination of the samples is a common problem.
We computed the following contrasts (in log2 fold changes) for each of the treatments (TS: TS mice; WT: wild type mice; NT: not treated mice; T, treated mice: whether TS.greentea, TS.EE, or TS.greentea + EE):
-
TS.NT − WT.NT (deregulated proteins in untreated TS mice).
-
TS.T − TS.NT (proteins responding to the treatments in TS mice).
-
WT.T − WT.NT (proteins responding to the treatments in WT mice).
-
(TS.T − TS.NT) − (WT.T − WT.NT) (interaction: proteins responding differently to the treatments in TS compared to WT mice).
For each protein/phosphopeptide whose abundance was significantly changing in the TS.NT − WT.NT we first calculated the log2-fold-change obtained in the contrast TS.NT − WT.NT (genotype gap). Thereafter, we computed the fold-change observed in the TS.T − WT.NT (treatment gap). We therefore calculated the fraction of recovered abundance, both in protein or phosphopeptides levels, after treatment—given by the difference between the genotype gap and the treatment gap—on the genotype gap: (genotype gap) – (treatment gap)/(genotype gap) (percentage of recovery).
When WT.NT is set to 0 (log2(1) = 0):
-
TS.NT − WT.NT can be written as TS.NT – log2(1) = TS.NT-0 = TS.NT.
-
TS.T − WT.NT is equal to the levels impaired at the basal state + levels after treatment = (TS.NT − WT.NT) + (TS.T − TS.NT) = TS.NT-0 + TS.T –TS.NT = TS.T.
And therefore:
(TS.NT − TS.T)/TS.NT = [(TS.NT − WT.NT) − (TS.NT − WT.NT) + (TS.T + TS.NT)]/(TS.NT − WT.NT).
The fraction of recovery could span from 0 (no recovery) to 1 (100% recovery). A value > 1 suggests overcorrection, while a value < 0 an impairment.
We defined as “rescued protein” the proteins, significantly changing (at the abundance or phosphorylation levels) inTS.NT-WT.NT, with a percentage of recovery from 50 to 150%; as “overcorrected proteins”, the ones with a percentage of recovery > 150; “not sufficiently rescued” those proteins with a percentage of recovery between − 50% and 50%; and “impaired proteins” those with a percentage of recovery < − 50%.
Similarly , we calculated the “percentage of impairment” by using in place of the TS.NT − WT.NT contrasts, its reverse WT.NT − TS.NT, to compute how much the treatment in the wild type was reducing the differences between TS and WT.
DYRK1A interactors
Given that previous work had identified EGCG as a potent DYRK1A inhibitor, we also explored DYRK1A interactors, taken from the mammalian verified interactors reported in previous literature6,34,35,36. We assessed the significance of the overlaps using a Fisher Exact Test.
Network analysis
To explore the functional significance of the differentially expressed and phosphorylated proteins, we built a protein–protein interaction network, in which the differentially abundant proteins and the proteins changing their phosphorylation status in TS vs WT were expanded to their direct interactors.
For the expansion, we used a list of bona fide physical interactors coming from the STRING datbase (version 10)37 and an internal database from Interactome3d version 2017_0138. We only considered interactions with a very high score (> 0.9) in STRINGdb and IMEx index39.
The visualization of the network was performed with the igraph R package40, using the Davidson Harel layout algorithm41. We used the ppi_enrichment function from the STRINGdb package42, to compute the odds of the observed number of the interaction compared to the expected ones.
To compute differences in the properties of the network between different sub-graphs of the network (i.e. differences in node degree) we permuted 1000 times the network, recalculating at each step the given properiesy on a random subset of the same size.
Enrichment analyses
We annotated our proteins with the R packages UniProt.ws and biomaRt43 (RRID:SCR_002987). We assessed the significance of the overlaps using a Fisher’s Exact test, considering as background the detected proteins specific for each contrast. We performed Gene Ontology Enrichment analysis with the clusterProfiler R package44 (RRID:SCR_016884).
Identification of significant hubs
We analyzed the heavy tail distribution of interactions with the poweRlaw R package45, comparing the power-law, Poisson, exponential, and log-normal distribution. We then used the fitted distribution for calculating the p-values associated with each protein corresponding to find by chance a higher number of interactions per protein than the given protein, setting the threshold to define hub at a p-value < 0.05.
Western blotting
Histones were isolated from whole hippocampi by acid-extraction from n animal, and for APP hippocampal proteins were extracted from 5 wild type and 5 Ts65Dn mice. After adding 6 × laemmli buffer to isolated histones or proteins stored in 2 M urea performed gel eletrophoresis and transferred the fractionated proteins to PVDF (only for histones) or nitrocellulose membranes using iBlot 2 (Thermofisher). The membranes were then blocked with 5% milk in TBS-Tween 0.1% (TBS-T) for 60 min before immunodetection with anti-diacetyl lysine 9 and lysine 14 histone H3 (AcH3K9K14) antibody (Cell Signaling Technology, #9677, RRID:AB_1147653) at a 1:1000 dilution, anti-histone H4 (Millipore 07-108, RRID:AB_2279758) at 1:500 dilution, anti-Actin at 1:2000 dilution (Sigma-Aldrich Cat# A2066, RRID:AB_476693), and anti-Amyloid Precursor Protein (Millipore Cat# AB5352, RRID:AB_91793) at 1:500 over-night at 4 ºC. Primary antibody incubation was followed by three washes (10 min, RT) in TBS-T before incubation with IRDye 800CW Goat anti-Rabbit IgG antibody (LI-COR Biosciences Cat# 925-32211, RRID:AB_2651127), three washes and visualization using Odissey Platform (LI-COR). Western blot’s bands were quantified by densitometry using ImageStudio.
Independent western blotting experiments were integrated by median normalization, excluding extreme values and statistical differences in the ratio AcH3/H4 or APP/ACTB were assessed with a linear mixed-effect model allowing for nested random effects (for sample coming from the same mouse).
Data availability
We deposited the mass spectrometry proteomics to the ProteomeXchange Consortium via the PRIDE46 partner repository (RRID:SCR_003411) with the dataset identifier EBI-PRIDE Accession PXD018515. We also created a repository with the R markdown file with the code to fully reproduce the analysis, the data showing that water intake was not altered when the treen tea extracts were dissolved in it, and the original images of the western blots: https://bitbucket.org/ilario_de_toma/proteomics_ts65dn.
References
de la Torre, R. et al. Safety and efficacy of cognitive training plus epigallocatechin-3-gallate in young adults with Down’s syndrome (TESDAD): a double-blind, randomised, placebo-controlled, phase 2 trial. Lancet Neurol. 15, 801–810. https://doi.org/10.1016/s1474-4422(16)30034-5 (2016).
De la Torre, R. et al. Epigallocatechin-3-gallate, a DYRK1A inhibitor, rescues cognitive deficits in Down syndrome mouse models and in humans. Mol. Nutr. Food Res. 58, 278–288. https://doi.org/10.1002/mnfr.201300325 (2014).
Takahashi, H. et al. Epigallocatechin-3-gallate increases autophagy signaling in resting and unloaded plantaris muscles but selectively suppresses autophagy protein abundance in reloaded muscles of aged rats. Exp. Gerontol. 92, 56–66. https://doi.org/10.1016/j.exger.2017.02.075 (2017).
Pervin, M. et al. Beneficial effects of green tea catechins on neurodegenerative diseases. Molecules https://doi.org/10.3390/molecules23061297 (2018).
Pons-Espinal, M., de Lagran, M. & Dierssen, M. Environmental enrichment rescues DYRK1A activity and hippocampal adult neurogenesis in TgDyrk1A. Neurobiol. Dis. 60, 18–31. https://doi.org/10.1016/j.nbd.2013.08.008 (2013).
Duchon, A. & Herault, Y. DYRK1A, a dosage-sensitive gene involved in neurodevelopmental disorders, is a target for drug development in down syndrome. Front. Behav. Neurosci. 10, 104. https://doi.org/10.3389/fnbeh.2016.00104 (2016).
De Toma, I., Ortega, M., Aloy, P., Sabidó, E. & Dierssen, M. DYRK1A overexpression alters cognition and neural-related proteomic pathways in the hippocampus that are rescued by green tea extract and/or environmental enrichment. Front. Mol. Neurosci. https://doi.org/10.3389/fnmol.2019.00272 (2019).
Martinez-Cue, C. et al. Differential effects of environmental enrichment on behavior and learning of male and female Ts65Dn mice, a model for Down syndrome. Behav. Brain Res. 134, 185–200. https://doi.org/10.1016/s0166-4328(02)00026-8 (2002).
Begenisic, T. et al. Environmental enrichment decreases GABAergic inhibition and improves cognitive abilities, synaptic plasticity, and visual functions in a mouse model of Down syndrome. Front. Cell. Neurosci. 5, 29. https://doi.org/10.3389/fncel.2011.00029 (2011).
Dierssen, M. et al. Alterations of neocortical pyramidal cell phenotype in the Ts65Dn mouse model of down syndrome: effects of environmental enrichment. Cereb. Cortex 13, 758–764. https://doi.org/10.1093/cercor/13.7.758 (2003).
Chakrabarti, L., Scafidi, J., Gallo, V. & Haydar, T. F. Environmental enrichment rescues postnatal neurogenesis defect in the male and female Ts65Dn mouse model of down syndrome. Dev. Neurosci. 33, 428–441. https://doi.org/10.1159/000329423 (2011).
Catuara-Solarz, S. et al. Principal component analysis of the effects of environmental enrichment and (-)-epigallocatechin-3-gallate on Age-associated learning deficits in a mouse model of down syndrome. Front. Behav. Neurosci. 9, 330. https://doi.org/10.3389/fnbeh.2015.00330 (2015).
Catuara-Solarz, S. et al. Combined treatment with environmental enrichment and (-)-epigallocatechin-3-gallate ameliorates learning deficits and hippocampal alterations in a mouse model of down syndrome. eNeuro https://doi.org/10.1523/ENEURO.0103-16.2016 (2016).
Reeves, R. H. et al. A mouse model for Down syndrome exhibits learning and behaviour deficits. Nat. Genet. 11, 177–184. https://doi.org/10.1038/ng1095-177 (1995).
Fernandez, F. et al. Normal protein composition of synapses in Ts65Dn mice, a mouse model of down syndrome. J. Neurochem. 110, 157–169. https://doi.org/10.1111/j.1471-4159.2009.06110.x (2009).
Peng, S., Zhang, Y., Zhang, J., Wang, H. & Ren, B. ERK in learning and memory: a review of recent research. Int. J. Mol. Sci. 11, 222–232. https://doi.org/10.3390/ijms11010222 (2010).
Tramutola, A. et al. Polyubiquitinylation profile in down syndrome brain before and after the development of alzheimer neuropathology. Antioxid. Redox Signal 26, 280–298. https://doi.org/10.1089/ars.2016.6686 (2017).
Lepri, F. et al. SOS1 mutations in Noonan syndrome: molecular spectrum, structural insights on pathogenic effects, and genotype-phenotype correlations. Hum. Mutat. 32, 760–772. https://doi.org/10.1002/humu.21492 (2011).
Bartsch, O. et al. DNA sequencing of CREBBP demonstrates mutations in 56% of patients with Rubinstein-Taybi syndrome (RSTS) and in another patient with incomplete RSTS. Hum. Genet. 117, 485–493. https://doi.org/10.1007/s00439-005-1331-y (2005).
Granno, S. et al. Downregulated Wnt/β-catenin signalling in the Down syndrome hippocampus. Sci. Rep. 9, 7322. https://doi.org/10.1038/s41598-019-43820-4 (2019).
Elias, G. M. & Nicoll, R. A. Synaptic trafficking of glutamate receptors by MAGUK scaffolding proteins. Trends Cell. Biol. 17, 343–352. https://doi.org/10.1016/j.tcb.2007.07.005 (2007).
de la Torre, R. & Dierssen, M. Therapeutic approaches in the improvement of cognitive performance in down syndrome: past, present, and future. Progress Brain Res. 197, 1–14. https://doi.org/10.1016/b978-0-444-54299-1.00001-7 (2012).
De Toma, I., Gil, L. M., Ossowski, S. & Dierssen, M. Where environment meets cognition: a focus on two developmental intellectual disability disorders. Neural Plast. 2016, 4235898. https://doi.org/10.1155/2016/4235898 (2016).
Peixoto, L. & Abel, T. The role of histone acetylation in memory formation and cognitive impairments. Neuropsychopharmacology 38, 62–76. https://doi.org/10.1038/npp.2012.86 (2013).
Fischer, A., Sananbenesi, F., Wang, X., Dobbin, M. & Tsai, L. H. Recovery of learning and memory is associated with chromatin remodelling. Nature 447, 178–182. https://doi.org/10.1038/nature05772 (2007).
Saldanha, S. N., Kala, R. & Tollefsbol, T. O. Molecular mechanisms for inhibition of colon cancer cells by combined epigenetic-modulating epigallocatechin gallate and sodium butyrate. Exp. Cell Res. 324, 40–53. https://doi.org/10.1016/j.yexcr.2014.01.024 (2014).
Thakur, V. S., Gupta, K. & Gupta, S. Green tea polyphenols increase p53 transcriptional activity and acetylation by suppressing class I histone deacetylases. Int. J. Oncol. 41, 353–361. https://doi.org/10.3892/ijo.2012.1449 (2012).
Fang, M. Z. et al. Tea polyphenol (-)-epigallocatechin-3-gallate inhibits DNA methyltransferase and reactivates methylation-silenced genes in cancer cell lines. Can. Res. 63, 7563–7570 (2003).
Haemisch, A. & Gärtner, K. Effects of cage enrichment on territorial aggression and stress physiology in male laboratory mice. Acta Physiol. Scand. Suppl. 640, 73–76 (1997).
Espadas, G., Borras, E., Chiva, C. & Sabido, E. Evaluation of different peptide fragmentation types and mass analyzers in data-dependent methods using an Orbitrap Fusion Lumos Tribrid mass spectrometer. Proteomics https://doi.org/10.1002/pmic.201600416 (2017).
Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26(12), 1367–1372 (2008).
Cox, J. et al. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805. https://doi.org/10.1021/pr101065j (2011).
Choi, M. et al. MSstats: an R package for statistical analysis of quantitative mass spectrometry-based proteomic experiments. Bioinformatics 30, 2524–2526. https://doi.org/10.1093/bioinformatics/btu305 (2014).
Aranda, S., Laguna, A. & de la Luna, S. DYRK family of protein kinases: evolutionary relationships, biochemical properties, and functional roles. FASEB J. 25, 449–462. https://doi.org/10.1096/fj.10-165837 (2011).
Guard, S. E. et al. The nuclear interactome of DYRK1A reveals a functional role in DNA damage repair. Sci. Rep. 9, 6539. https://doi.org/10.1038/s41598-019-42990-5 (2019).
Roewenstrunk, J. et al. A comprehensive proteomics-based interaction screen that links DYRK1A to RNF169 and to the DNA damage response. Sci. Rep. 9, 6014. https://doi.org/10.1038/s41598-019-42445-x (2019).
Szklarczyk, D. et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447-452. https://doi.org/10.1093/nar/gku1003 (2015).
Mosca, R., Ceol, A. & Aloy, P. Interactome3D: adding structural details to protein networks. Nat. Methods 10, 47–53. https://doi.org/10.1038/nmeth.2289 (2013).
Orchard, S. et al. Protein interaction data curation: the international molecular exchange (IMEx) consortium. Nat. Methods 9, 345–350. https://doi.org/10.1038/nmeth.1931 (2012).
Csardi, G. & Nepusz, T. The igraph software package for complex network research. Int. J. Compl. Syst. 1, 1695 (2005).
Harel, D. & Koren, Y. A Fast Multi-Scale Method for Drawing Large Graphs Vol. 6 (Springer, New York, 2000).
Pradines, J. R., Farutin, V., Rowley, S. & Dancik, V. Analyzing protein lists with large networks: edge-count probabilities in random graphs with given expected degrees. J. Comput. Biol. 12, 113–128. https://doi.org/10.1089/cmb.2005.12.113 (2005).
Durinck, S., Spellman, P. T., Birney, E. & Huber, W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 4, 1184–1191. https://doi.org/10.1038/nprot.2009.97 (2009).
Yu, G., Wang, L.-G., Han, Y. & He, Q.-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287. https://doi.org/10.1089/omi.2011.0118 (2012).
Gillespie, C. Fitting Heavy Tailed Distributions: The poweRlaw Package. R package version 64, https://doi.org/10.18637/jss.v064.i02 (2014).
Vizcaino, J. A. et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, D447-456. https://doi.org/10.1093/nar/gkv1145 (2016).
Acknowledgements
This work was supported by the Fondation Jérôme Lejeune Foundation (Project #1347 “Elucidation of the mechanism of action of epigallocatechin-3-gallate as a therapeutic agent on the cognitive phenotype in Down Syndrome mice models”), Agencia Estatal de Investigación (AEI) (PID2019-110755RB-I00/AEI/10.13039/501100011033), H2020 SC1 Gene overdosage and comorbidities during the early lifetime in Down Syndrome GO-DS21-848077, Fundació La Marató De TV3 (201620-31_MDierssen), EU (JPND HEROES (tHE cRossroad Of dEmentia Syndromes), NIH (Grant Number: 1R01EB 028159-01). The CRG is a Center of Excellence Severo Ochoa. The CIBER of Rare Diseases is an initiative of the ISCIII. The laboratories of M. Dierssen and E. Sabidó are supported by DIUE de la Generalitat de Catalunya (Grups consolidats 2017SGR926, 2017SGR595). The CRG/UPF Proteomics Unit is part of the Spanish Infrastructure for Omics Technologies (ICTS OmicsTech) and it is a member of the ProteoRed PRB3 consortium which is supported by grant PT17/0019 of the PE I+D+i 2013-2016 from the Instituto de Salud Carlos III (ISCIII) and ERDF. We also acknowledge the support of the Spanish Ministry of Science and Innovation to the EMBL partnership, the Centro de Excelencia Severo Ochoa and the CERCA Programme/Generalitat de Catalunya.
Author information
Authors and Affiliations
Contributions
I.D. performed the bioinformatic analyses, the APP western blot, and wrote the manuscript. M.O. performed the mass proteomic experiment and some preliminary analyses. S.C. reared and treated the mice. C.S. performed the western blot for the global histone acetylation levels. M.D. and E.S. conceived the project and M.D. wrote and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
De Toma, I., Ortega, M., Catuara-Solarz, S. et al. Re-establishment of the epigenetic state and rescue of kinome deregulation in Ts65Dn mice upon treatment with green tea extract and environmental enrichment. Sci Rep 10, 16023 (2020). https://doi.org/10.1038/s41598-020-72625-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-72625-z
This article is cited by
-
The lncRNA Snhg11, a new candidate contributing to neurogenesis, plasticity, and memory deficits in Down syndrome
Molecular Psychiatry (2024)
-
Pharmacological Inhibition of p-21 Activated Kinase (PAK) Restores Impaired Neurite Outgrowth and Remodeling in a Cellular Model of Down Syndrome
Neurotoxicity Research (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.