Abstract
The mechanisms by which the human cerebral cortex folds into its final form remain poorly understood. With most of the current models and evidence addressing secondary folds, we sought to focus on the global geometry of the mature brain by studying its most distinctive feature, the Sylvian fissure. A digital human fetal brain atlas was developed using previously obtained MRI imaging of 81 healthy fetuses between gestational ages 21 and 38 weeks. To account for the development of the Sylvian fissure, we compared the growth of the frontotemporal opercula over the insular cortex and compared the transcriptome of the developing cortices for both regions. Spatiotemporal mapping of the lateral hemispheric surface showed the highest rate of organized growth in regions bordering the Sylvian fissure of the frontal, parietal and temporal lobes. Volumetric changes were first observed in the posterior aspect of the fissure moving anteriorly to the frontal lobe and laterally in the direction of the temporal pole. The insular region, delineated by the limiting insular gyri, expanded to a much lesser degree. The gene expression profile, before folding begins in the maturing brain, was significantly different in the developing opercular cortex compared to the insula. The Sylvian fissure forms by the relative overgrowth of the frontal and temporal lobes over the insula, corresponding to domains of highly expressed transcription factors involved in neuroepithelial cell differentiation.
Similar content being viewed by others
Introduction
The human cerebral cortex at birth has completed the formation of all major gyral and sulcal folds. The cerebral surface grows from a largely smooth surface beginning at the sixth month of gestation to the characteristic patterns of gyrification1,2,3,4,5,6,7,8,9,10. A number of hypotheses have been proposed on the mechanisms of preterm brain folding11,12. These include mechanical instability that can arise from the outer gray matter expanding at a faster rate than the underlying white matter, the axonal tension hypothesis in which white matter axons draw together overlying cortical regions to form gyri, and genetic prepatterning of the cortex to form convolutions2,3,13,14. Postnatally, cortical expansion continues through differential local growth that extend into adulthood, without altering the newborn’s brain folded outline15. With most of the current evidence focusing on the formation of intralobar folds, how the brain acquires its final global form is largely unknown, including the development of its most distinctive feature, the formation of the Sylvian fissure2,3,5,13,16,17,18.
Observations of cortical development in preterm infants have shown that maximal directional growth occurs from the central sulcus toward the parietal lobe, then toward the frontal and temporal regions19,20. The normal process of cortical development follows a predictable sequence21. The Sylvian fissure, the deepest sulcus on the lateral hemispheric surface, can be identified as early as 12 weeks of gestation and serves as a major landmark for the dynamic changes of the brain surface. Abnormal morphologic features of the Sylvian fissure can be frequently associated with neuronal migration disorders22,23. The development of this prominent fold defines the global shape of the brain and cannot be explained by current models, which render aleatory sulci and gyri, with no distinctive and reproducible large scale structure18.
The Sylvian fissure forms by the convergence of the frontal and temporal lobes over the insula, which is a distinctive and unique mechanism not shared by any other sulcus24. The characteristic radial migration pathway from the ventricular and subventricular zones, that forms most of the brain gyri, appears to be absent in the insula, which cells originate from the pallial/subpallial boundary and migrate in an oblique fashion around the basal ganglia25.
In the present study, our goal is to evaluate the development of the Sylvian fissure as a result of asymmetric cortical growth between the frontoparietal and temporal opercula with respect to the insular cortex. Our hypothesis is that the convergence of the frontal and temporal opercula over the insula, driven by discrete regions of high growth, are responsible for the formation of the fissure and are associated with differential genetic expression patterns.
Results
Differential cortical expansion and convergence drives the formation of the Sylvian fissure
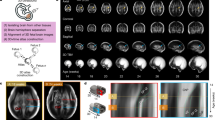
We calculated the local expansion (growth) of each area of the brain by registering each gestational week to the next one on a week by week basis. To determine local growth, we calculated the Jacobian determinant, which represents the local volumetric change (Fig. 1). This spatiotemporal mapping of the lateral hemispheres demonstrates early growth in the frontal, temporal, and parietal opercula, closing the Sylvian fissure from gestational weeks (GW) 23–25. Our analysis demonstrates that focal areas of cortical growth (volumetric expansion) and convergent growth of the opercula close the Sylvian fissure. In later weeks (GW 27–31) differential cortical expansion drives the formation of the major cortical sulci, progressing outward from the central sulcus in the frontal and parietal lobes and inferiorly in the temporal lobe. Although comparable, the left and right hemispheres exhibited asymmetric deformations and gained volume in similar places but at slightly different times, never exceeding 2 weeks of dyscoordination.
Volumetric changes on the lateral hemispheric surface during gestation. Spatiotemporal mapping of the lateral hemispheres. Blue–red color code indicates the Jacobian determinant of the week-to-week co-registration warping, a measure of local volume growth. The Jacobian determinant is normalized for global volume growth, as described in the methods. Postconceptional weeks are indicated. As can be observed, the highest rate of organized growth localizes to the opercula of insula. The first of these “hot zones” is located to the supramarginal gyrus, with the frontal and temporal poles increasing their local expansion at later times. The scale represents the weekly proportional growth (i.e. 1.1 represents a 10% weekly relative expansion). Note that week labels are gestational week (GW).
Using the same registration described previously, we created a vector map demonstrating the magnitude and direction of local tissue displacements (Fig. 2). This analysis demonstrates the direction of tissue growth in contrast to the Jacobian analysis which demonstrates the degree of overall tissue growth. Only cortical regions exhibited high magnitude vectors whereas the telencephalic white matter rendered neutral or small deformation levels. Both the temporal and frontotemporal opercula converged over the insula, coinciding with volumetric growth described previously. Convergence of the frontal and temporal lobes over the insula led to the formation of the Sylvian fissure. Notably, no posteroventral folding or bending was observed. This was evident at every gestational age but diminished in magnitude as the gestation progressed (Fig. 2).
Vector displacement during development. Week-to-week displacements, calculating local tissue displacement, after normalizing for volumetric growth, were calculated for each voxel. The magnitude (total displacement) is indicated by the white to red scale (black is shading from arrows) Axial, median and coronal views at the indicated postconceptional weeks are shown. High magnitude vectors are observed close to the evolving cortex but are sparse in the white matter. The frontoparietal and temporal opercula converge over the insular cortex that is ultimately covered by them forming the Sylvian fissure. The opercula correspond to areas of high relative growth consistently throughout gestation. Note that week labels are gestational week (GW).
Developmental gene expression
Cortical progenitors commonly form and migrate outward from the ventricular zone by using radial glial fibers as a scaffold. As a consequence, neurons born from the same progenitor area in the ventricular zone occupy neighboring positions in the mature cortex. This is, however, not true for the insular cortex, which neurons migrate obliquely from the pallial/subpallial boundary around the basal ganglia25. We therefore analyzed the transcriptomes of the maturing cortices of the areas of high growth (defined by the opercula outside the insular limiting sulci, target structure) and the insula (limited by the limiting insular sulci, contrast structure). We report the top 0.1% most overexpressed genes relative to the insula, for each cellular layer of interest: subpial granular zone (SZ), marginal zone (MZ), cortical plate (CP) and subcortical plate (SP) (Table 1).
We found extensive differences in the transcriptome for every region, which may indicate a different origin of the cells forming these structures and may drive their differing growth rates. In the subpial granular zone, the 0.1% relative highest expressed genes had a 13.7-fold (opercula/insula) expression ratio (Fig. 3A). Amongst these, the SH3YL1 gene (related to cellular migration) and the SOCS7 gene (related to hydrocephalus in mice) are involved in CNS development. In the marginal zone the cutoff was 8.2-fold (Fig. 3B). Among the genes overexpressed in the opercular regions related to brain growth, we found DIO3, which deactivates T4 to T3 conversion; NTF3, involved in the formation of thalamocortical connections and neurite growth and survival and IRX2, involved in developmental pattern formation. For the cortical plate the fold difference to reach the 0.1 percentile was 3.4 (Fig. 3C). Several of the most overexpressed genes in this region are involved in neuronal growth including SNAI2, related to epithelial mesenchymal transitions; ATXN3, related to transcriptional regulation and neuroprotection; GREM1, proposed to control development by selectively antagonizing the activities of the transforming growth factor (TGF)-beta ligand; NTF3; GABRA4, that regulates GABAergic activity and; EZR4, that acts as a scaffold between the actin cytoskeleton and transmembrane proteins facilitating cell–cell interactions and receptor retention and DOCK7, that plays a role in priming neurites to become the axons. For the subcortical plate the threshold for reporting was 6.9 (Fig. 3D) including IRX2 and NEUROD1, that generates functional neurons from human pluripotent stem cells.
Comparison of the genetic expression in the maturing cortex of the opercula and the insula at 21 post-conceptional weeks (GW 23). A marked difference between the transcriptome of cortical areas (frontal parietal, temporal opercula) and the insular cortex. This may drive the higher degree of expansion that was observed at every level of the maturing cortex [(A) subpial granular zone; (B) marginal zone; (C) cortical plate and (D) subplate]. M1 primary motor cortex, S1 primary somatosensory cortex, Lateral T/O lateral temporooccipital. Ref: 2010 Allen Institute for Brain Science. BrainSpan Atlas of the Developing Human Brain51.
Discussion
During the last two-thirds of gestation, the cerebral cortex expands and folds into a conserved arrangement of sulci and gyri1,2,3,5. The physical mechanisms by which the distinctive convoluted cortical pattern develops have been addressed in numerous publications18,19,20. Nevertheless, these models fail to account for the global geometry of the brain and do not explain the development of the Sylvian fissure18,19,20. Here, we found that the anterior and posterior poles of the telencephalic vesicle converge over the central region forming the frontal and temporal lobes, and the Insula, respectively. This process is triggered by the differential expansion of the fronto-temporal opercula very early during GW 23–25, preceding any bending process22. These two different regions exhibited a consistent difference in their transcriptomes in the pre-folded brain, with the temporal, frontal and parietal opercular cortices having consistent expression patterns that were different from that of the insula.
We interrogated the BrainSpan Atlas to determine which genes are preferentially expressed in the developing cortices of the frontal, parietal and temporal lobes compared to the insular at 21 PCW, corresponding to gestation week (GW) 23. Given the lack of longitudinal data in the BrainSpan dataset, we were not able to determine the temporal sequence of transcriptional changes that lead to folding nor the specific pathways by which these transcriptional changes effect changes in growth. Per our analysis of the Gholipour atlas, the Sylvian fissure largely closes from 23 to 25 GW, suggesting that genetic expression immediately preceding this may drive this process. Further, while MRI changes may lag anatomic changes, the closure of the Sylvian fissure is a large-scale process that is clearly visible on fetal MRI (Fig. 1). Our aim was to identify transcriptional differences at the beginning of this period (week 21) that may drive this difference in growth pattern.
Many of the overexpressed genes in the opercula have a well-defined or plausible role in neuronal migration or survival during development. SOCS7 is a protein coding gene that regulates signaling cascades and its involved in terminating neuronal migration26. Although the process is complex and incompletely understood, it is known that the complex SOCS7-Cul5-Rbx2 regulates termination of migration and disruption of that system results in abnormally position neurons in the CP26.
Similarly, Neurotrophin 3 (NTF3), IRX2, and DOCK7, have roles in neuronal differentiation and survival. NTF3 controls the survival and differentiation of neurons and is thought to promote neuronal survival in the developing brain27,28. In contrast, while IRX2 does not have a defined role in telencephalic development, it is known to participate in the rostro caudal differentiation of the hindbrain29. DOCK7 codes for a guanine nucleotide exchange factor, which in turn activates Rac 1 and 3 and Rho amongst others.DOCK7 has a direct role in brain development by regulating the fate of the radial glia30,31. Mutations on this gene cause epileptic encephalopathy and cortical blindness32.
Other proteins we identified do not have a clearly identified role (as of yet) in brain development. Ataxin 3 (ATX3) is involved in protein ubiquitination, a trinucleotide expansion of this gene (CAG) causes Machado-Joseph disease or spinocerebellar ataxia, although this mutation is associated with neuronal death, so far ATX does not have a well-defined role in brain development33,34. GREM1 codes for a bone morphogenetic protein (BMP) antagonist and can modulate organogenesis. In mouse it conveys the Sonic Hedgehog polarizing signal and is involved in limb development. A role in cortical folding has not been described35.
Apart from the physical principles involved in brain folding, numerous reports evidenced that this process is under heavy genetic regulation5,36,37,38,39,40. Recently de Juan Romero et al. described a genetic patterning in the ferret’s VZ and outer ventricular zone (OVZ), matching the folded geometry of the brain and present before gyrification started. These differentially expressed genes (DEG) are composed of thousands of sequences that are expressed in an on and off fashion alternating between sulci and gyri, including Trnp1, Ccnd1, EOMES, Notch, Shh, MAPK and Wnt17,39,40. This pattern conforms a blueprint for individual folds but do not explain the large-scale arrangement of the brain lobes.
A comparison between our findings and the DEGs described by de Juan Romero would be inaccurate; on one side they analyzed the ventricular and subventricular zones and not the cortices themselves; on the other, the mechanism involved in forming cortical convolutions in ferrets and humans are different and may be governed by different mechanisms39,41,42. The genetic expression at the pallial/subpallial boundary was described by Carney et al.43. This region is highly heterogeneous and expresses a mixed array of genes from the pallium (Pax6) and subpallium (Gsh2).
Our findings suggest that the insular progenitor cells have a distinct transcriptional signature than the populations that develop into the frontal/temporal/parietal operculae. The latter expand at a much higher rate than the former, thus closing the Sylvian fissure on the lateral hemispheric surface. The work of de Juan Romero et al.40 and Carney et al.43 suggests that the insular progenitors may follow an oblique migration pathway in contrast to the direct radial migration pathway leading to the lateral hemispheric cortical surface (Fig. 4). The developing basal ganglia may block a direct pathway for progenitor cells in the ventricular zone to the insula. The distinct gyral configuration of the insula (5 relatively linear gyri that converge to the limen insulae) in contrast to the convoluted gyri of the lateral hemispheric surface may also be related to this44. Future studies must investigate the genetic makeup of these progenitor populations and how this drives the connectivity and morphology of the insula and operculae.
Different origins and migration paths explain the differences in growth between the insula and the frontal, parietal and temporal lobes and define the large-scale morphology of the human brain. Schematic of a 21 post-conceptional week (GW 23) brain. Radial migration from the ventricular zone (blue arrows) forms almost the entire lateral hemispheric cortical surface (blue) and is linked to the appearance of known sulci and gyri. On the other hand, the insular cortex (green) is isolated from periventricular proliferating zones by the basal ganglia and is formed by cells originating from the pallial-sub pallial boundary, defining an oblique migration pathway, which is linked with less expansion and morphologically distinct sulci.
The atlas developed by Gholipour et al. is derived from 81 healthy, normally-developing fetuses45, while the gene expression data is derived from two 21 PCW (23 GW) fetuses as described in the BrainSpan documentation. It is important to note that the Gholipour atlas uses gestational week as a measure of time, which is approximately 2 weeks ahead of post-conceptional week. Thus, the genetic information from the BrainSpan Atlas corresponds to the very beginning of the Gholipour atlas. Although different individuals were used to construct both datasets, we argue that the genetic drivers of Sylvian fissure formation are highly conserved across individuals given the universal formation of the fissure in the healthy human brain.
Our findings are limited by the small number of analyzed specimens, the use of bulk mRNA instead of single cell expression, and the difficulty of matching the time and spatial dimensions between a dataset generated from MRI and one constructed directly from fetal brain tissue. Given the ethical constraints in obtaining fetal brain tissue, the number of specimens available for genetic analysis is inherently limited. Further, given these same ethical limits, longitudinal imaging and genetic analysis is not possible in the same individual. Despite these constraints, this study deepens our understanding of the geometry and the transcriptome of the normal development of the Sylvian fissure.
Conclusion
The Sylvian fissure forms by the relative overgrowth of the frontal and temporal lobes over the insula, with the developing cortices exhibiting sharply different transcriptomes. It is interesting to note that some of these genes are related to neuronal proliferation and differentiation functions and can be part of the landscape of the genes responsible for the general geometry of the brain.
Methods
Guidelines statement
All data used in this analysis were obtained from publicly available databases that were obtained in accordance with all relevant guidelines and regulations. Specifically, for the fetal MRI atlas by Gholipour et al.45 the study was approved by the Boston Children’s Hospital Institutional Review Board and the Committee on Clinical Investigation and written informed consent was obtained from all participants. For the BrainSpan atlas, all work was performed following guidelines for the research use of human brain tissue of the University of Washington and Advanced Bioscience Resources (Alameda, California) with approval by the Human Investigation Committees and Institutional Ethics Committees of each institute.
Developmental MRI assessment
Atlas
We analyzed the fetal brain atlas described by Gholipour et al.45,46. This atlas was reconstructed from weekly fetal T2-weighted MRI images of 81 healthy fetuses from gestational week 21 to 38 segmented to identify multiple key subcortical and cortical regions. A description of the Atlas and its full content can be accessed at https://crl.med.harvard.edu/research/fetal_brain_atlas/.
Registration
To analyze fetal brain development over time, we utilized the SyN algorithm in the ANTs (Advanced Normalization Tools)47 package to register the atlas fetal MRI on a week by week basis. This is a nonlinear, diffeomorphic registration paradigm. We performed two types of registration. First (part A), we registered images on a week-by-week basis (week 38 to week 37, week 37 to week 36, etc.) using the full SyN algorithm in the reverse direction.
Second (Part B), in order identify nonlinear changes such as gyral growth or folding and ignore gross volumetric changes, we registered all weeks to week 38 using an affine transformation (performed using ANTs). Subsequently we performed nonlinear registration using SyN to identify these nonlinear changes.
Registration validation
In order to validate the accuracy of our registration, we warped the week 38 segmentation to the desired week using our registration method45. We then compared the overlap of the atlas segmentation for the desired week to the warped segmentation using the multiclass Dice similarity coefficient, as implemented in the Scikit-learn package in Python.
Jacobian analysis
To determine volumetric changes on a week by week basis, we calculated the Jacobian determinant of Part A registration described above (week-to-week) using ANTS. The Jacobian determinant is the determinant of the Jacobian matrix, a matrix of the first order partial derivatives of the transformation, and represents the local volumetric change associated with the transformation. A determinant greater than 1 represents expansion and vice versa. We used this to generate Jacobian maps for each week representing the relative volume expansion/contraction for each voxel for each week (after correcting for global scaling). Note that the Jacobian determinants are normalized for global volume change—e.g. if overall global volume growth was 9%, an area with Jacobian determinant of 0.95 still grew 3.5%. We rendered these maps in 2D and 3D using Nibabel, matplotlib, and nilearn, all publicly available Python packages48,49.
Displacement maps
To visualize the direction and magnitude of local displacements, we rendered the normalized (part B), registration using arrow plots, which allow for visualization of local deformation50. In addition, we overlaid these plots with a heat map of the magnitude (total displacement) of each area. This was performed using Nibabel and matplotlib50.
Developmental gene expression
Atlas
In the current study, the BrainSpan atlas was utilized to profile gene transcription at different stages of brain maturation. The BrainSpan atlas is an ARRA-funded grant through the NIH to a consortium consisting of the Allen Institute for Brain Science; Yale University; the University of Southern California; Massachusetts General Hospital, Harvard-MIT Health Sciences and Technology, Athinoula A. Martinos Center for Biomedical Imaging; the University of California, Los Angeles; and the University of Texas Southwestern Medical Center51. All data are publicly accessible. The methods and processes used to generate gene expression data have been previously published and can be accessed via https://www.brainspan.org51.
Postmortem human brain specimens were from the following: the Department of Neurobiology at Yale School of Medicine and the National Institute of Mental Health; the Human Fetal Tissue Repository at the Albert Einstein College of Medicine; the Brain and Tissue Bank for Developmental Disorders at the University of Maryland; the Birth Defects Research Laboratory at the University of Washington; the MRC-Wellcome Trust Human Developmental Biology Resource at the Institute of Human Genetics, University of Newcastle, U.K. The protocol was approved by the respective IRBs. Informed consent was obtained.
The BrainSpan Atlas sampled over 90 substructures in the developing brain, listed at https://help.brain-map.org/download/attachments/3506181/Prenatal_LMD_Microarray.pdf, building on the previous work by Kang et al.7. This includes the granular and dysgranular insular cortices (listed under “Neocortex” in Table 2 in the white paper at the link), and the surrounding opercular areas.
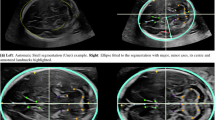
Because the BrainSpan Atlas transcriptome is not referenced in the stereotactic space (it is provided in coronal slices), we defined the insula and operculae with anatomical landmarks visible both in the MRI dataset and the BrainSpan Atlas. The insula was defined as the cortical region within the limiting insular sulci, which corresponded to the dysgranular and granular insular cortices in the BrainSpan Atlas. We then identified substructures in the BrainSpan atlas that correspond to the superior (frontal and parietal lobe), and inferior (temporal) operculae. Note that not all substructures had transcriptomic data for all layers (subpial granular zone, marginal zone, cortical plate, subplate). The included opercular structures from the BrainSpan atlas were—frontopolar cortex, ventrolateral prefrontal cortex, orbital frontal cortex, primary motor cortex, primary somatosensory cortex, inferior parietal cortex, parainsular temporal cortex (e.g. auditory cortex), lateral temporooccipital cortex, and superior temporal cortex. We acknowledge that several of these structures span areas beyond the conventional definition of the operculae but given the parcellation from the BrainSpan atlas, we were limited to this. This is illustrated in Figs. 5 and 6.
Matching areas of relatively high and slow growth with their respective transcriptomes. In order to match anatomical areas on the MR with defined regions on the transcriptome reference atlas, we referenced our analyses to constant anatomical landmarks present in both. Based on the results rendered by the MR analysis, the area within the boundaries of the limiting insular gyri, with a low relative growth rate, was compared with the fast growing opercular cortices. From superficial to deep the developing cortical regions analyzed were the subpial granular zone (SG), the marginal zone (MZ), cortical plate (CP) and the subplate (SP). The dashed blue lines connect the insular limiting sulci, which were used as reference, since they are clearly demarcated both in the genomics atlas (A) as well as present in our MR analysis (B).
Locations of prenatal microarray profiling at PCW 21 (GW 23) and demarcations of cortical areas. Per the methodology described at the Allen Brain Atlas (https://help.brain-map.org/download/attachments/3506181/Prenatal_LMD_Microarray.pdf), the insula and surrounding operculae were specifically delineated and profiled. This figure highlights these areas—specifically for the insula (blue) and the frontal-parietal (superior—yellow) and temporal (inferior—green) operculae. The included structures from the BrainSpan atlas were—granular insular cortex, dysgranular insular cortex, frontopolar cortex, ventrolateral prefrontal cortex, orbital frontal cortex, primary motor cortex, primary somatosensory cortex, inferior parietal cortex, parainsular temporal cortex (e.g. auditory cortex), lateral temporooccipital cortex, and superior temporal cortex. (A) Surface illustration of the insula and operculae at PCW 21 (GW 23). The red squares indicate the locations of slices. (B) Coronal slices at indicated locations. These coronal slices are directly comparable to the BrainSpan atlas sections. Each region was sampled at as many as four layers—SG subpial granular zone, MZ marginal zone, CP cortical plate, SP subplate.
Comparative whole transcriptome analysis
We analyzed the transcriptional profile at 21 postconceptional weeks (PCW) available for a total of two specimens. We interrogated the atlas based on the result of the fetal MR analysis described above. We aimed to compare the expression profile of the developing cortex in areas of high growth with cortical regions exhibiting minimal relative expansion. The subpial granular zone (SZ), marginal zone (MZ), cortical plate (CP) and subcortical plate (SP) corresponding to the frontoparietal and temporal opercula were compared to the insula.
A comparative search of genes consistently over expressed in the target regions (opercular cortex) compared to the contrast region (insular cortex) was performed. We report on the 99.9 highest expressed percentile of around 35,000 genes analyzed for each region, which selects the top 35 overexpressed genes in each area. The cut off (in folds) was 13.7 for the SG, 8.2 for the MZ, 3.4 for the CP and 6.9 for the SP.
References
Chi, J. G., Dooling, E. C. & Gilles, F. H. Gyral development of the human brain. Ann. Neurol. 1, 86–93 (1977).
Bayly, P. V., Taber, L. A. & Kroenke, C. D. Mechanical forces in cerebral cortical folding: A review of measurements and models. J. Mech. Behav. Biomed. Mater. 29, 568–581 (2014).
Tallinen, T. et al. On the growth and form of cortical convolutions. Nat. Phys. 12, 588–593 (2016).
Garel, C. et al. Fetal cerebral cortex: Normal gestational landmarks identified using prenatal MR imaging. Am. J. Neuroradiol. 22, 184–189 (2001).
Fernández, V., Llinares-Benadero, C. & Borrell, V. Cerebral cortex expansion and folding: What have we learned?. EMBO J. 35, 1021–1044 (2016).
Bystron, I., Blakemore, C. & Rakic, P. Development of the human cerebral cortex: Boulder Committee revisited. Nat. Rev. Neurosci. 9, 110–122 (2008).
Kang, H. J. et al. Spatio-temporal transcriptome of the human brain. Nature 478, 483–489 (2011).
Paredes, M. F. et al. Extensive migration of young neurons into the infant human frontal lobe. Science 354, 6308 (2016).
Piao, X. et al. G protein-coupled receptor-dependent development of human frontal cortex. Science 303, 2033–2036 (2004).
Zilles, K., Armstrong, E., Schleicher, A. & Kretschmann, H. J. The human pattern of gyrification in the cerebral cortex. Anat. Embryol. (Berl.) 179, 173–179 (1988).
Van Essen, D. C. A tension-based theory of morphogenesis and compact wiring in the central nervous system. Nature 385, 313–318 (1997).
Zilles, K. et al. Quantitative analysis of sulci in the human cerebral cortex: Development, regional heterogeneity, gender difference, asymmetry, intersubject variability and cortical architecture. Hum. Brain Mapp. 5, 218–221 (1997).
Tallinen, T. & Biggins, J. S. Mechanics of invagination and folding: Hybridized instabilities when one soft tissue grows on another. Phys. Rev. E Stat. Nonlinear Soft Matter Phys. 92, 2 (2015).
Vasung, L. et al. Quantitative and qualitative analysis of transient fetal compartments during prenatal human brain development. Front. Neuroanat. 10, 11 (2016).
Li, G. et al. Mapping region-specific longitudinal cortical surface expansion from birth to 2 years of age. Cereb. Cortex 23, 2724–2733 (2013).
Garcia, K. E., Kroenke, C. D. & Bayly, P. V. Mechanics of cortical folding: Stress, growth and stability. Philos. Trans. R. Society B Biol. Sci. 373, 1759 (2018).
Borrell, V. How cells fold the cerebral cortex. J. Neurosci. 38, 776–783 (2018).
Tallinen, T., Chung, J. Y., Biggins, J. S. & Mahadevan, L. Gyrification from constrained cortical expansion. Proc. Natl. Acad. Sci. 111, 12667–12672 (2014).
Hill, J. et al. Similar patterns of cortical expansion during human development and evolution. Proc. Natl. Acad. Sci. USA 107, 13135–13140 (2010).
Garcia, K. E. et al. Dynamic patterns of cortical expansion during folding of the preterm human brain. Proc. Natl. Acad. Sci. USA 115, 3156–3161 (2018).
Serag, A. et al. Construction of a consistent high-definition spatio-temporal atlas of the developing brain using adaptive kernel regression. Neuroimage 59, 2255–2265 (2012).
Sarnat, H. B. & Flores-Sarnat, L. Telencephalic flexure and malformations of the lateral cerebral (Sylvian) fissure. Pediatr. Neurol. 63, 23–38 (2016).
Mochida, G. H. Molecular genetics of lissencephaly and microcephaly. Brain Nerve 60, 437–444 (2008).
Bush, A., Nuñez, M., Brisbin, A. K., Friedlander, R. M. & Goldschmidt, E. Spatial convergence of distant cortical regions during folding explains why arteries do not cross the sylvian fissure. J. Neurosurg. 1–10 (2019). (online ahead of print)
González-Arnay, E., González-Gómez, M. & Meyer, G. A radial glia fascicle leads principal neurons from the pallial-subpallial boundary into the developing human insula. Front. Neuroanat. 11, 111 (2017). https://doi.org/10.3389/fnana.2017.00111.
Sekine, K., Kubo, K. I. & Nakajima, K. How does Reelin control neuronal migration and layer formation in the developing mammalian neocortex?. Neurosci. Res. 86, 50–58 (2014).
Anderson, K. D. et al. Differential distribution of exogenous BDNF, NGF, and NT-3 in the brain corresponds to the relative abundance and distribution of high-affinity and low-affinity neurotrophin receptors. J. Comp. Neurol. 357, 296–317 (1995).
Rodríguez-Tébar, A., Dechant, G. & Barde, Y. A. Neurotrophins: Structural relatedness and receptor interactions. Philos. Trans. R. Society Lond. Series B Biol. Sci. 331, 255–258 (1991).
Nakamura, H., Katahira, T., Matsunaga, E. & Sato, T. Isthmus organizer for midbrain and hindbrain development. Brain Res. Rev. 49, 120–126 (2005).
Yamauchi, J., Miyamoto, Y., Chan, J. R. & Tanoue, A. ErbB2 directly activates the exchange factor Dock7 to promote Schwann cell migration. J. Cell Biol. 181, 351–365 (2008).
Gadea, G. & Blangy, A. Dock-family exchange factors in cell migration and disease. Eur. J. Cell Biol. 93, 466–477 (2014).
Perrault, I. et al. Mutations in DOCK7 in individuals with epileptic encephalopathy and cortical blindness. Am. J. Hum. Genet. 94, 891–897 (2014).
Johnson, S. L. et al. Differential toxicity of ataxin-3 isoforms in drosophila models of spinocerebellar ataxia type 3. Neurobiol. Dis. 132, 104535 (2019).
Nóbrega, C. et al. Molecular mechanisms and cellular pathways implicated in Machado-Joseph disease pathogenesis. Adv. Exp. Med. Biol. 1049, 349–367 (2018).
Johnson, E. J., Neely, D. M., Dunn, I. C. & Davey, M. G. Direct functional consequences of ZRS enhancer mutation combine with secondary long range SHH signalling effects to cause preaxial polydactyly. Dev. Biol. 392, 209–220 (2014).
Lohmann, G., Von Cramon, D. Y. & Colchester, A. C. F. Deep sulcal landmarks provide an organizing framework for human cortical folding. Cereb. Cortex 18, 1415–1420 (2008).
Walsh, C. A. Genetic malformations of the human cerebral cortex. Neuron 23, 19–29 (1999).
Lohmann, G., von Cramon, D. Y. & Steinmetz, H. Sulcal variability of twins. Cereb. Cortex 9, 754–763 (1999).
Reillo, I. & Borrell, V. Germinal zones in the developing cerebral cortex of ferret: Ontogeny, cell cycle kinetics, and diversity of progenitors. Cereb. Cortex 22, 2039–2054 (2012).
de Juan Romero, C., Bruder, C., Tomasello, U., Sanz-Anquela, J. M. & Borrell, V. Discrete domains of gene expression in germinal layers distinguish the development of gyrencephaly. EMBO J. 34, 1859–1874 (2015).
Elsen, G. E. et al. The protomap is propagated to cortical plate neurons through an Eomes-dependent intermediate map. Proc. Natl. Acad. Sci. USA 110, 4081–4086 (2013).
Baala, L. et al. Homozygous silencing of T-box transcription factor EOMES leads to microcephaly with polymicrogyria and corpus callosum agenesis. Nat. Genet. 39, 454–456 (2007).
Carney, R. S. E., Cocas, L. A., Hirata, T., Mansfield, K. & Corbin, J. G. Differential regulation of telencephalic pallial-subpallial boundary patterning by Pax6 and Gsh2. Cereb. Cortex 19, 745–759 (2009).
Türe, U., Yaşargil, D. C. H., Al-Mefty, O. & Yaşargil, M. G. Topographic anatomy of the insular region. J. Neurosurg. 90, 720–733 (1999).
Gholipour, A. et al. A normative spatiotemporal MRI atlas of the fetal brain for automatic segmentation and analysis of early brain growth. Sci. Rep. 7, 476 (2017).
Marami, B. et al. Temporal slice registration and robust diffusion-tensor reconstruction for improved fetal brain structural connectivity analysis. Neuroimage 156, 475–488 (2017).
Avants, B. B. et al. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage 54, 2033–2044 (2011).
Tustison, N. J. & Avants, B. B. Explicit B-spline regularization in diffeomorphic image registration. Front. Neuroinform. 7, 39 (2013).
Avants, B. B., Epstein, C. L., Grossman, M. & Gee, J. C. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med. Image Anal. 12, 26–41 (2008).
Venthur, B., Dähne, S., Höhne, J., Heller, H. & Blankertz, B. Wyrm: A brain–computer interface toolbox in python. Neuroinformatics 13, 471–486 (2015).
Miller, J. A. et al. Transcriptional landscape of the prenatal human brain. Nature 508, 199–206 (2014).
Funding
This research was partially supported by the American Association of Neurological Surgeons via the Van Wagenen Fellowship.
Author information
Authors and Affiliations
Contributions
E.G., A.B. and A.N.M. were involved in the design and conception of this manuscript. E.G., H.D. and A.M. performed the literature search. E.G., A.B., H.D. and A.N.M. compiled the primary manuscript. E.G., A.B., A.K.B. and A.M. compiled the figures. E.G., A.M. and A.B. critically revised the manuscript. All authors have approved the manuscript as it is written. E.G., A.M., A.K.B., H.D. all contributed to figures and tables. All figures/tables were created or assembled by the co-authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mallela, A.N., Deng, H., Brisbin, A.K. et al. Sylvian fissure development is linked to differential genetic expression in the pre-folded brain. Sci Rep 10, 14489 (2020). https://doi.org/10.1038/s41598-020-71535-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-71535-4
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.