Abstract
The human microbiome plays a key role in maintaining host homeostasis and is influenced by age, geography, diet, and other factors. Traditionally, India has an established convention of extended family arrangements wherein three or more generations, bound by genetic relatedness, stay in the same household. In the present study, we have utilized this unique family arrangement to understand the association of age with the microbiome. We characterized stool, oral and skin microbiome of 54 healthy individuals from six joint families by 16S rRNA gene-based metagenomics. In total, 69 (1.03%), 293 (2.68%) and 190 (8.66%) differentially abundant OTUs were detected across three generations in the gut, skin and oral microbiome, respectively. Age-associated changes in the gut and oral microbiome of patrilineal families showed positive correlations in the abundance of phyla Proteobacteria and Fusobacteria, respectively. Genera Treponema and Fusobacterium showed a positive correlation with age while Granulicatella and Streptococcus showed a negative correlation with age in the oral microbiome. Members of genus Prevotella illustrated high abundance and prevalence as a core OTUs in the gut and oral microbiome. In conclusion, this study highlights that precise and perceptible association of age with microbiome can be drawn when other causal factors are kept constant.
Similar content being viewed by others
Introduction
Aging is an extremely complex, perpetual, progressive and multifactorial process resulting in decreased physiologic functions of all the organ systems1. Studies have reported that the human microbiome, the latest discovered organ, is also significantly influenced by increasing age2,3,4. Studies have suggested that the complex and diverse communities of microbes that inhabit the gut vary through different stages of an individual’s life5. Many of these alterations are harmless and natural, while some of the alterations can have an important effect on overall homeostasis6. Importantly, notable changes in the human microbiome occur when the immune system is relatively weak, i.e., at the start of life and during aged life7. Many of the fluctuations in the microbiome are harmless and natural; nonetheless, studies have shown that some disturbances in the gut microbiome can have important effects on health and disease6. Earlier studies on human microbiome analyses have discovered an increased abundance of Bacteroides species in the elderly population8. An enriched abundance of Proteobacteria a bacterial group containing ‘pathobionts’ known for causing impairment in a susceptible host, i.e., in centenarian’s2,3,4. But the populations used for these studies were either from different geographical locations or with different diets2,3,4. In agreement with the studies on the human gut microbiome, in older micro-pigs9 also decreased abundance of beneficial microbes including probiotic bacteria and short-chain fatty acid-producers was recorded while Bacteroides were increased with the increasing age9.
Several studies have shown age-associated effects in the experimental animals wherein, transfer of an aged microbiome into germ-free mice leads to systemic inflammation10,11, on the contrary replacement of microbiome of the aged mice with the microbiome of younger mice boosts the local germinal center reactions1. Researchers noted that the aged mice’s germinal center reaction was stronger following fecal transplantation from the younger mice1. Moreover, remodeling of the gut microbiome has shown to increase the lifespan in diverse organisms, including Drosophila melanogaster12,13, killifish14, mice15, etc.
Coevolved with the host and its ancestors for millions of years, the microbiome plays a key role in the maintenance of host health and wellbeing by performing various functions ranging from digestion, protection against pathogen colonization to host immunity and regulation of central nervous16,17,18,19,20,21. Also, the microbiome is responsive to a variety of confounding factors such as host ethnicity22,23, age24,25, diet26,27 and geographic location24,28. Hence, it is imperative to study the same population longitudinally for the exploration of a precise association of age and microbiome. Studying the microbiome of genetically linked individuals of multiple generation families having similar diet, ethnicity and staying in the same geographic regions can help in estimating the perseverance of microbiota obtained in early years and its progression with age. Traditionally, India has an established convention of the joint family system, which is an extended family arrangement consisting of three or more generations living in the same household structure bound by genetic relatedness29,30. Especially in rural and semi-urban joint families in India, family members live in the same house structure and have similar dietary and sanitary habits. Hence, the Indian population provides a unique opportunity to understand age-related changes in the human microbiome. Studies relating to the influence of FUT2 and birth mode variants on microbiome31, diabetes-associated microbiome32, obesity-related microbiome33, the microbiome of celiac disease patients34, an association of microbiome with ayurvedic Prakriti types35, microbiome structure of rural, urban36 and tribal37 populations were carried out in the Indian populations. However, age-related changes in the human microbiome across different body habitats are unknown. The majority of studies understanding the age and human microbiome correlations have been focused on the gut microbiome, while changes in oral and skin microbiome in the aging process have been relatively less studied.
In the present study, we provide a comprehensive analysis of the human microbiome from the gut, oral and skin ecosystems from 54 healthy subjects belonging to six different patrilineally related three generation families staying together in rural Indian settings. The study population has harmonized dietary, social habits, hygiene and sanitation habits, economic status and geographic position. Predominantly, other microbiome contributing factors were harmonized and age was the only distinguishing factor. 16S rRNA gene amplicon sequencing-based microbiome analysis performed to investigate the age-related changes in the gut, skin and oral microbiome of Endogamous Agriculturist Indian (EAI) sub-population.
Results
Gut, oral and skin microbiome profile of EAI population
Overall, a total of 9,566,497 sequences were generated, out of which 8,048,975 (File S1) were taxonomically assigned, resulting in 6,708 OTUs for the gut microbiome. Four samples (St.D1004, St.D301, St.S610 and St.S612) were removed from further analysis due to lower sequencing depth. Bacterial phyla such as Bacteroidetes (49.3%), Firmicutes (41.6%), Proteobacteria (5.7%), Actinobacteria (2.18%) and Tenericutes (0.4%) were highly dominant and constituted ~99% of the total gut microbiome. Presence of 174 different bacterial genera was noted, wherein Prevotella (50%), Dialister (12%), Bacteroides (9%), Megamonas (3%), and Succinivibrio (3%) were among the dominant taxa contributing to a total of 77% of the gut microbiome (Fig. S1a). Although, genus Prevotella was observed to have varying relative abundance ranging from 2% to 77% across the study population, its dominance was evident from the fact that 62% (n = 31) of the study subjects had an abundance in a range of 33% to 77% of the total gut microbiome (Table S1).
In the oral microbiome, 7,568,649 good quality sequences (File S1) clustered into 2,167 OTUs. Analysis based on the taxonomic assignment of these reads revealed a higher abundance of bacterial phyla such as Proteobacteria (34%), Bacteroidetes (32%), Firmicutes (24%), Fusobacteria (6%) and Actinobacteria (2%) constituting 96% of the oral microbiome. Genera Neisseria (20%), Streptococcus (15%), Prevotella (14%), Porphyromonas (10%), and Haemophilus (10%) were found to be the five most dominant genera totaling up to 69% of the oral microbiome (Fig. S1b). The relative abundance of genus Prevotella was observed to be more than 10% in half of the population (Table S2).
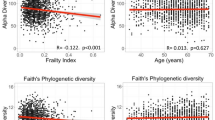
The skin microbiome data comprised of 10,951,175 good quality sequences (File S1) clustered into 10,920 OTUs. Skin microbiome analysis showed a higher abundance of phyla Firmicutes (49%), Proteobacteria (26%), Actinobacteria (12%) and Bacteroidetes (8%) collectively on 11 different body sites including dry, moist and sebaceous regions. Only one percent of OTUs were assigned to phylum Cyanobacteria. The skin microbiome showed the highest number of OTUs (n = 10,920) compared to oral and stool samples. Corynebacterium (10%) Alloiococcus (9%), Staphylococcus (8%), Streptococcus (7%) and Anaerococcus (6%) were the most dominant and diverse bacterial genera detected in the skin microbiome (Fig. S1c). Alpha diversity analysis measures, i.e., observed species (OTUs), Chao1, Shannon and Inverse Simpson revealed no significant differences in the gut and skin microbiome when compared between the three age groups (Fig. 1), family structure and dietary habits (Table 1). However, significant differences were observed in the oral microbiome between the age groups (ANOVA, p < 0.05 with Benjamini-Hochberg FDR corrections) (see Fig. 1B).
Contribution of core taxa in the gut, oral and skin microbiome of patrilineal families
Bacterial genera prevalent in 95% of the study population with more than 0.1% abundance were considered as a part of the core microbiome. Estimation of core microbiome was performed for individual families (n = 6 families) and the overall EAI study population. Amongst the 171 total bacterial genera detected in the gut microbiome, only three genera, namely Prevotella, Ruminococcus and Faecalibacterium, were recorded as a part of core microbiome across all the families (Fig. 2a, Table S3). These core taxa represented 23% to 91% gut microbiome composition of the participants (Fig. S2). With the aforementioned detection threshold few bacterial genera were explicitly detected in particular family as core taxa wherein, Parabacteroides was detected in family D3, Haemophilus and Roseburia in family D8, Streptococcus and Dorea in family D10 (Fig. S3, Table S3).
The core microbiome of oral samples represented presence of 13 (6.7%) bacterial genera amongst the 192 total genera. These genera include Neisseria, Streptococcus, Prevotella, Porphyromonas, Haemophilus, Fusobacterium, Granulicatella, Veillonella, Capnocytophaga, Rothia, Aggregatibacter, Gemella and Lautropia (Fig. 2b). Overall, these core taxa estimated 79% to 96% of the total microbiome composition (Fig. S2).
In the skin microbiome samples, Corynebacterium and Streptococcus were the only bacterial genera detected as core taxa across all the families (Fig. 2c, Table S5). Similar to the gut microbiome, family-specific bacterial genera were also detected in the skin microbiome. These genera include Novosphingobium in family D8, Enhydrobacter, Salinicoccus and Butyrivibrio in family D10, and Haemophilus and Gemella in family S6 (Fig. S5, Table S5). These core taxa represented 37% to 94% of the total microbiome (Fig. S2). Largely, the contribution of core taxa in the gut, oral and skin microbiome of all the patrilineal families was identical.
Influence of diet on the gut microbiome
Detailed dietary information of the study population was collected using the food frequency questionnaire (FFQ). With the help of a nutritionist, the dietary information was subsequently converted into the daily intake of carbohydrates, proteins, fats, lipids, fibers and calories (Table S6). Investigation revealed that average carbohydrates consumption in the first, second and third-generation members was 166, 396 and 339 grams, providing 74%, 81% and 80% of daily calories in the respective generations. Overall the type and amount of dietary components were similar across the population, except for family D3, which has relatively lesser consumption of these components. Canonical correspondence analysis (CCA) based on the abundance of bacterial genera, amount of dietary components and samples metadata showed that all the samples were scattered across the ordination plot and no clear clustering of the samples was observed based on age group or gender (Fig. 3). The variation explained by the ordination plot was also non-significant, reporting 7.5% for the gut, 10.6% for the oral and 6.9% for the skin microbiome (Fig. 3). Also, correlation analysis between the relative abundance of bacterial taxa and routine consumption of dietary components showed no significant association (Fig. 4).
Association of age and microbiome
Microbiome community structure of gut, oral and skin samples was investigated across three generations (age groups). Amongst the prevalent bacterial genera of the gut microbiome, Succinivibrio and Ruminococcus were highly abundant in the age group 1, Dialister, Megamonas, Phascolarctobacterium, Megasphaera and Faecalibacterium in the age group 2 and Prevotella, Bacteroides and Bifidobacterium were in the age group 3 (Table S7a). Likewise, in the oral microbiome Prevotella, Fusobacterium, Veillonella, Capnocytophaga, Rothia and Aggregatibacter were highly abundant in the age group 1, genus Haemophilus in age group 2 while Neisseria, Streptococcus, Porphyromonas and Granulicatella in the age group 3 (Table S7b). High abundance of few bacterial taxa was recorded in particular age groups in the skin microbiome samples also, wherein Corynebacterium, Alloiococcus, Peptoniphilus, Haemophilus, Acinetobacter and Clostridium were highly abundant in age group 1, Anaerococcus, Porphyromonas and Campylobacter in age group 2 and Staphylococcus, Streptococcus, Novosphingobium, Paracoccus, Moraxella and Prevotella in age group 3 (Table S7c). Primarily, age-associated changes were observed in the microbiome structure of three-generation members and to strengthen these observations; statistical analysis was also completed. Comparative microbiome analysis in three age groups showed no significant differential abundance of bacterial genera in the gut and skin microbiome. However, the oral microbiome showed significant variations in the abundance of genera Dialister, Fusobacterium, Streptococcus, Selenomonas, Filifactor and Treponema (Fig. 5D) (ANOVA, p < 0.05 with Benjamini-Hochberg FDR corrections). We confirmed our observations using qPCR analysis for quantifying the absolute proportion of genus Prevotella in the total human gut bacteria (Fig. 5E).
Nonmetric Multidimensional Scaling (NMDS) ordination displaying microbiome communities across the three generations in the gut (A), oral (B) and skin (C) microbiome. (D) Box plot showing differentially abundant genera in the human oral microbiome across the members from the three age groups. (E) Next-generation sequencing and qPCR results showing the abundance of Prevotella and total bacteria in the human gut microbiome across three age groups.
Beta diversity analysis using non-metric multidimensional scaling (NMDS) plots based on Bray-Curtis metrics showed no clear clustering in the samples based on the age groups of the study population (Fig. S6). Age-associated changes in the microbiome were further analyzed based on differentially abundant OTUs (ANOVA, p < 0.05 with Benjamini-Hochberg FDR corrections). Investigation revealed the presence of 69 (1.03%) differentially abundant OTUs across three age groups in the gut microbiome. Similarly, 190 (8.66%) and 293 (2.68%) differentially abundant OTUs were observed in human oral and skin microbiome, respectively. A high number of differentially abundant OTUs were present in the oral samples. Beta diversity analysis using these differentially abundant OTUs showed clustering of samples based on the age groups in the gut, oral and skin samples (Fig. 5A–C).
We further performed a correlation analysis of gut, oral and skin microbiome with age. Linear regression analysis using the nonparametric Spearman correlation revealed a higher abundance of phylum Proteobacteria with increasing age in the gut microbiome (p < 0.05) (Fig. 6A). While, in the oral microbiome of the population, a higher abundance of phylum Fusobacteria was observed with the increasing age (p < 0.05) (Fig. 6B). However, no such age-based correlations were observed in the skin microbiome.
Amidst the total 171 bacterial genera in the gut microbiome, only genus Bacteroides showed age-associated changes. Decreased abundance Bacteroides was recorded with the increasing age (nonparametric Spearman correlation (p < 0.05) (Fig. 6B). Whereas, in the oral microbiome of the population, bacterial genera Treponema and Fusobacterium showed a positive correlation (Fig. 6D,E) while genera Granulicatella and Streptococcus showed a negative correlation with the age (p < 0.05) (Fig. 6F,G). However, in the skin microbiome, no such statistically significant correlations were noted.
Discussion
Indian patrilineal extended family structure provides a unique opportunity to study the underlying effects of age, diet, and genetics influencing the human microbiome. Such family structure is a widely seen residential unit comprising of 2–4 patrilineally related generations living together, particularly in rural and semi-urban settings29,30. In the present study, we analyzed gut, oral and skin microbiome from 54 healthy subjects belonging to six different families from a single biogeographic region (Dongargaon: 18.6199° N, 74.0807° E and Shikrapur: 18.6924° N, 74.1323° E).
In the gut and oral microbiome, we found a high prevalence of Prevotella (Fig. 2a,b; Tables S1 and S2), a bacterial genus known to be associated with degradation of complex plant polysaccharides38,39. Indian diet is rich in plant-based carbohydrates, and our observations are consistent with earlier reports where a high prevalence of Prevotella in the gut microbiome of the Indian population was observed39,40. Prevalence of Prevotella has also been reported in the African population consuming a diet rich in carbohydrates and fibers41.
Understanding the confounding factors that shape and define the oral microbiome is crucial for understanding the broader systemic health42, as oral microbiome has long been known to be a reservoir for infection at other body sites43. In our analysis, high abundance of bacterial genera Neisseria, Streptococcus, and Prevotella were observed (Fig. S1b). Genus Neisseria is aerobic and primary colonizers of the oral cavity, Streptococcus is a facultative anaerobe while Prevotella is the obligate anaerobic bacteria. The high abundance of aerobic, facultative anaerobic and obligate anaerobic bacteria suggests the role of oxygen sensitivity in structuring the composition of bacterial diversity associated with the oral cavity. This diverse microbiome can perform versatile metabolic functions crucial for the healthy oral cavity. Earlier, these bacterial genera reported being common residents of the oral cavity in different populations in the healthy state44,45,46,47.
Investigating influencing factors is important to determine homeostatic forces that contribute to a healthy skin microbial community48. In the skin microbiome, Corynebacterium, Alloiococcus, and Staphylococcus were found to be the most abundant genera (Fig. S1c). These observations are in agreement with those reported by earlier studies in diverse ethnic groups, globally49. In contrast with an earlier study that reported Actinobacteria as the most dominant phylum of the skin microbiome50, we observed dominance of Firmicutes followed by Proteobacteria and then Actinobacteria. These differences in skin microbiome of EAI sub-population could be associated with unique genetics, ethnicity and environmental conditions.
Further, different regions of the human skin like dry, moist and sebaceous are known to harbor different microbial community51. These differences could not be ascertained in our study since skin samples collected from 11 different body sites were pooled together before sequencing. The skin of healthy individuals generally harbors low microbial biomass and it requires sufficient starting material52. Hence DNA from all the 11 body locations of the participants were extracted separately and eventually pooled together before sequencing to avoid sequence artifacts associated with low biomass samples52.
Microbial diversity, which contributes to the core microbiome, can provide a snapshot of homeostasis in the population and deviations from this core can be associated with different physiological states53. Presence of three, 13 and two core genera were observed in the gut, oral, and skin microbiome, respectively (Fig. 2). Genus Dialister was also amongst the highly abundant core taxa of the gut microbiome in all the families of the EAI population (Figs. 2, S3). An earlier study has reported that microbial enzymatic repertoire is known for the conversion of dietary fibers into short-chain fatty acids (SCFA)54. Our observations of a higher abundance of Dialister in Indian sub-population along with high consumption of dietary fiber suggests a need to test this possible association, as these bacteria are previously reported for SCFA production (propionate)55. Gut and oral microbiome showed a high proportion of core microbiome compared to the skin microbiome. Amongst the core taxa in oral microbiome, Porphyromonas are known for the expression of the fimA gene, which encodes for the surface protein important for attachment to other oral bacteria56. The oral microbial flora comprises diverse human-associated biofilms, which are influenced by oral streptococci, the main group of early colonizers57. Fusobacterium was also prevalent taxa in the oral microbiome. In the complex ecosystems like oral cavity microbial co-aggregations like Fusobacterium and Streptococcus, which mediates a variety of cooperative metabolic functions57. Skin is the largest organ and represents the primary physical barrier between the host and the external environment. The overall representation of the core microbiome was only 15%, presumably due to oil, moist and sebaceous site-specific bacterial community structure and transient nature of the skin microbiome. Due to the acidic pH of the skin (4.4 to 5), despite the transient nature of skin microbiome, only mutualistic skin bacteria like Streptococcus, Staphylococcus and Corynebacterium can grow and detected as core taxa. Unique combination of taxa such as Dialister, Prevotella, Bacteroides, Megamonas, Succinivibrio in gut, Streptococcus, Fusobateria Neisseria, Prevotella, Porphyromonas in oral and Corynebacterium, Alloiococcus, Staphylococcus, Streptococcus, Anaerococcus and Peptoniphilus in skin were observed in EAI subpopulation emphasizing the effect of diet, host genetics and environmental factors on microbiome. Overall the core microbiome structure of the EAI population was similar across all the families which can be associated with similar dietary patterns, socioeconomic status, ethnicity, and agriculture-based lifestyle. Few low abundant bacterial taxa were exclusively detected as core taxa of specific families (Tables S3–S5). However, no relatable information was observed for the distinctive presence of these taxa.
Dietary information of the study population was collected using the food frequency questionnaire (FFQ) and this information is subsequently translated into the daily intake of carbohydrates, proteins, fats, lipids, fibers and calories with the help of a nutritionist. Detailed analysis showed that carbohydrates provide about 74%, 81% and 80% calories in the first, second and third generation members, respectively (Table S6) and overall, the consumption of other dietary constituents was comparable across the three generations. Correlation analysis of bacterial genera and routine dietary consumption of carbohydrates, proteins, fats, lipids, fibers and calories showed that there is no statistically significant correlation suggesting the relatively similar structure of microbiome and overall dietary pattern (Fig. 4). This observation is further strengthened by limited variation observed in the CCA analysis and no specific clustering was observed based on the generation (age groups) or the gender of the study participants (Fig. 3). A balanced diet helps in maintaining human health and the changes in the diet are responsible for the associated alterations in the microbiome. Singh et al., have shown that dietary alterations can induce microbiome associated changes in 24 hours, which can be alternating and yet reproducible58. Patrilineal families in this study follow the typical diet for several generations, and generally, all members of the family eat the same food irrespective of their age. The routine diet of the study populations comprises majorly of wheat and/or pearl millet bread, rice, vegetables and millets. The correlation analysis on this population revealed no statistically significant differences in the microbiome and the diet. This emphasizes the fact that overall homogeneity in the diet helps in maintaining the microbial state. Other confounding factors, including birth mode (cesarean section delivery and normal delivery), monozygotic or dizygotic twins had no effect on the microbiome as all the participants recruited in the study have the same normal delivery birth mode and none of the participants were twins.
A substantial number of studies have reported the association between age and the human microbiome59,60, but the majority of these studies were among unrelated individuals who lacked constant causal contributing factors. Participants from three generations belonging to patrilineal families and living in the same household were recruited in this study to understand the perceptible effect of age on the microbiome. This sampling strategy allowed us to have a minimum impact of other confounding factors on the microbiome. Human microbiome dynamics changes with the time as the ‘holobiont’ integrates and responds to signals from the environment61. A direct causal relation between age-specific microbial communities and host aging has also been explored in laboratory model organisms, including flies, fish and mice1,12,13,14,15,16, etc. Microbiome community structure of gut, oral and skin samples illustrated differences in the abundance of bacterial genera in three age groups. Here, Succinivibrio known for higher fiber degrading potential62 and Ruminococcus were highly abundant in first-generation members (Table S7a). The specific reason for the higher abundance of these taxa is not known and it demands further investigation. Bacterial taxa known for healthier metabolism were abundant in the gut microbiome of the second generation members such as Dialister and Phascolarctobacterium the SCFA producers63,64, Megasphaera the key carbohydrate metabolizing bacteria of Indian population known for having diverse and unique sets of Carbohydrate-Active enzymes (CAZymes)65, Faecalibacterium is also the most abundant and important commensal bacteria of the human gut microbiota8. In addition to Prevotella, Bifidobacterium, the early gut colonizers and Bacteroides were higher in the third generation members (Table S7a). In the skin microbiome also age-related changes in the abundance of bacterial taxa were recorded. Genus Corynebacterium was highly abundant in first-generation members (Table S7c). Recent study understanding the extrinsic and intrinsic host factors influencing skin microbiome composition suggested that Corynebacterium OTUs were associated with skin aging66, specifically with the hyperpigmented spots and wrinkles66. With the increasing age, physiological changes occur in the skin structure explains the association of key bacterial taxa in the members of the respective age groups. In our study, only 1.03% OTUs were found to be differentially abundant across three age groups, suggesting a nominal but profound effect of age on the gut microbiome. With the increasing age, the high abundance of Proteobacteria was detected (Fig. 6A). A higher abundance of this bacterial phylum was reported to be associated with the altered gut microbiome and dysbiosis67,68. Studies have shown an increase in the abundance of Proteobacteria with age correlating with the weaker immune response to the opportunistic pathogens, thereby leading to a decrease in the commensal microflora69,70. Proteobacteria have been suggested as the potential diagnostic criteria for dysbiotic conditions7.
Similarly, in the oral microbiome, Fusobacteria was found to increase with increasing age (Fig. 6C). Few genera of this phylum are known opportunistic pathogens71; however, studies on the association of members of this phylum longitudinally with age can give more insights into their mutualistic or pathogenic role. Further, we observed a negative correlation in the abundance of Bacteroides with age (Fig. 6B); this is in contrast to previous studies demonstrating the higher abundance of genus Bacteroides with increasing age72,73. With the increasing age, physiological changes occur in the oral cavity like thinning of oral mucosa, smooth and loosened stippling aspect, narrowing and alteration of the gingival epithelium, modification of epithelial-connective interface and decreasing of keratinization74. Here, Granulicatella and Streptococcus abundance decreased with age (Fig. 6D,E) while Treponema and Fusobacterium abundance increased with age (Fig. 6F,G). Granulicatella is the component of normal oral flora and Streptococcus is also normal flora and early colonizers of the oral microbial community. On the contrary, few members of the genera Treponema and Fusobacterium are opportunistic pathogens. These age-related changes could be associated with the physiological changes in the oral cavity with the increasing age.
This study expressly describes the age-related changes in the microbiome. However, analysis of hematological and biochemical parameters of blood may have further provided an opportunity to understand its association with the microbiome, the clear picture on age-related changes in the overall metabolism and health and disease status. Further studies with additional samples and multoimics approach can help strengthen these findings.
In conclusion, this study particularly highlights the precise and perceptible association of age with the microbiome. Our finding suggests that the age-related changes are very specific and bacterial phylum Proteobacteria needs to be investigated in detail to understand its specific physiological role in gut microbiome. Similarly, bacterial taxa, including Treponema, Fusobacterium, Granulicatella and Streptococcus the member of the human oral microbiome, can be explored for their importance in the oral microbiome. Also, the findings suggest that core taxa constitute more than 75% of the gut and oral microbiome, while only 67% of the skin microbiome, indicating a larger variability of the microbiome present on the skin. We present baseline data of the human microbiome from a healthy Indian sub-population, which could be used as a reference for further studies, including diabetes75,76,77 obesity and inflammatory diseases.
Methods
Ethical clearance declaration
The study was approved by the ethics committees of the National Centre for Cell Science (NCCS), Pune and King Edward’s Memorial Hospital Research Centre (KEMHRC), Pune. Written informed consent from the study subjects or their parents wherever applicable were taken, as per the guidelines of the institutional ethics committee and Indian Council of Medical Research (ICMR), India. We confirm that all the experiments were performed as per the approved guidelines.
Recruitment of subjects
Subjects were recruited from the Vadu Health and Demographic Surveillance System (Vadu HDSS) area of the Vadu Rural Health Program, KEM Hospital Research Centre, Pune (VRHP, KEMHRC, Pune). The Vadu study population comprises of about 170,000 individuals that reside in 22 villages. The objective of Vadu HDSS is to create a longitudinal database of demographic information, including fertility, mortality, migration and marital status, of the Vadu area. Two villages, namely Dongargaon (Latitude: 18.7442326, Longitude: 73.4504317) and Shikrapur (Latitude: 18.687639, Longitude: 74.125671) were selected out of the total Vadu HDSS region.
The recruitment was done based on the following criteria.
-
1.
Minimum three generations (I- age >50 years, II- age between 25 to 40 years and III- age between 3 to 15 years) with at least two members per generation must be living together in the same house structure.
-
2.
Self-declared healthy individuals.
-
3.
Families with individuals having a history of consumption of alcohol, tobacco and recent (last 3 months) use of antibiotics were excluded from the study.
Among the 30 families screened, six families comprising of 54 individuals fulfilled the required criteria and were included in the study.
Metadata and sample collection
A Food Frequency Questionnaire (FFQ), along with 48 hrs dietary recall was administered before sample collection. Detailed information on the consumption of the food item and quantity for each meal of the day were recorded. With the help of nutritionists, this information then translated into the daily consumption of carbohydrates, proteins, lipids, fibers and calories. Additional metadata about the use of antibiotics and medicines, hygiene and sanitary practices, lifestyle, socioeconomic status, social habits, health and diseases (self-reported with or without medical records based on standard questionnaire) and other demographic characteristics were recorded. Separate health status questionnaires for adults and children were administered for the selection of healthy adults for the study (Tables S8 and S9).
Detailed information on routine dietary consumption of different food nutriments, their frequency and quantity were collected and recorded from the study participants. This information was recorded for three important meals, i.e., breakfast, lunch, and dinner. Also, the data on routine consumption of any additional specific food nutriment besides these three meals was recorded. The information on routine dietary consumption was then used for calculating the routine consumption of carbohydrates, proteins, lipids, fibers, and calories.
Gut, oral, and skin samples from the recruited subjects were collected in triplicates (with an interval of one week). Freshly voided, early morning fecal sample was collected in a sterile container. Early morning oral washing (before brushing or gargling) was collected using freshly prepared sterile 1X PBS (pH 7.4) in a sterile container. Skin samples from 11 different body sites per individual (belonging to three different regions, i.e., moist, oily and sebaceous region) were collected as described in Fig. S7. All samples were stored at −80 °C until further processing.
Microbiome profiling
DNA extraction from fecal (representative of the gut), oral and skin samples was done using QIAamp stool DNA mini kit, QIAamp DNA mini kit, and QIAamp blood and tissue DNA extraction kit, respectively (Qiagen, USA). The DNA extraction was performed according to the manufacturer’s instruction with the inclusion of bead beating and freeze-thaw treatment at −80 °C and 90 °C for 10 minutes alternatively. Metagenomic sequencing of the V3-V4 region of the 16S rRNA gene was done using Illumina Miseq platform, paired-end (2 × 300 bp) sequencing, as described earlier78.
Bioinformatics and statistical analysis
Assembly of paired-end reads for each sample was carried out using FLASH (Fast Length Adjustment of SHort reads). Low-quality sequences were removed during the assembly with low overlapping regions (less than 20 nucleotides)79. Microbial diversity analysis was done using standard QIIME (v1.8.0) pipeline80. Closed reference-based OTU picking approach was used to cluster reads into Operational Taxonomic Units (OTUs) at 97% sequence similarity using UCLUST algorithm81 and Greengenes database (13.8) and representative sequences from each OTU were selected for taxonomic assignment. Beta diversity and other statistical analysis was performed using Phyloseq82, corrplot83, vegan84 and Microbiome85 packages in R. Additional statistical analysis were performed using STAMP86 and GraphPad Prism (GraphPad Software, La Jolla California USA). A web-based tool InteractiVenn was used for the analysis of shared and unique bacterial genera87.
Quantification of genus Prevotella in study population
Quantitation of genus Prevotella and total bacteria from fecal samples was carried out using qPCR as described previously88. Briefly, for quantifying 16S rRNA gene for total bacteria and Prevotella, 10 μl reactions in triplicate were set containing a suitable pair of primers88 (Table 2), 50 ng of Metagenomic DNA and SYBR green master mix (Applied Biosystems Inc. USA), using 7300 Real-time PCR system (Applied Biosystems Inc. USA). Following PCR conditions: initial denaturation at 95 °C for 10 min, followed by 40 cycles at 95 °C for 10 s, 60 °C for 1 min was used. Group-specific standard curves were generated from serial dilutions of a known concentration of respective PCR products. Additionally, melting curve analysis was performed at the end of qPCR cycles to check the amplification specificity. Average values of the triplicate were used for enumerations of tested gene copy numbers for each group using standard curves.
Data availability
The sequence data is available at NCBI SRA submission with accession number SRP116277 (Bioproject ID: PRJNA399246) for gut microbiome, SRP135853 (Bioproject ID: PRJNA438584) for skin microbiome and SRP135913 (Bioproject ID: PRJNA438728) for oral microbiome.
References
Stebegg, M. et al. Heterochronic faecal transplantation boosts gut germinal centres in aged mice. Nature Communications 10 (2019).
Biagi, E. et al. Through Ageing, and Beyond: Gut Microbiota and Inflammatory Status in Seniors and Centenarians. PLoS One 5, e10667 (2010).
Round, J. L. & Mazmanian, S. K. The gut microbiota shapes intestinal immune responses during health and disease. Nature Reviews Immunology 9, 313–323 (2009).
Claesson, M. J. et al. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proceedings of the National Academy of Sciences 108, 4586–4591 (2010).
Thursby, E. & Juge, N. Introduction to the human gut microbiota. Biochemical Journal 474, 1823–1836 (2017).
Ravinder Nagpal et al. Gut microbiome and aging: Physiological and mechanistic insights. NHA 4, 267–285 (2018).
Arrieta, M.-C., Stiemsma, L. T., Amenyogbe, N., Brown, E. M. & Finlay, B. The Intestinal Microbiome in Early Life: Health and Disease. Frontiers in Immunology 5 (2014).
Rinninella, E. et al. What is the Healthy Gut Microbiota Composition? A Changing Ecosystem across Age, Environment, Diet, and Diseases. Microorganisms 7, 14 (2019).
Lim, M. Y., Song, E.-J., Kang, K. S. & Nam, Y.-D. Age-related compositional and functional changes in micro-pig gut microbiome. GeroScience (2019).
Fransen, F. et al. Aged Gut Microbiota Contributes to Systemical Inflammaging after Transfer to Germ-Free Mice. Frontiers in Immunology 8 (2017).
Thevaranjan, N. et al. Age-Associated Microbial Dysbiosis Promotes Intestinal Permeability, Systemic Inflammation, and Macrophage Dysfunction. Cell Host & Microbe 21, 455–466.e4 (2017).
Obata, F., Fons, C. O. & Gould, A. P. Early-life exposure to low-dose oxidants can increase longevity via microbiome remodelling in Drosophila. Nature Communications 9 (2018).
Westfall, S., Lomis, N. & Prakash, S. Longevity extension in Drosophila through gut-brain communication. Scientific Reports 8 (2018).
Smith, P. et al. Regulation of life span by the gut microbiota in the short-lived African turquoise killifish. eLife 6 (2017).
Sonowal, R. et al. Indoles from commensal bacteria extend healthspan. Proceedings of the National Academy of Sciences 114, E7506–E7515 (2017).
Huttenhower, C. et al. Structure, function and diversity of the healthy human microbiome. Nature. 486(7402), 207–214, https://doi.org/10.1038/nature11234 (2012).
Qin, J. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 (2010).
Wang, J. & Jia, H. Metagenome-wide association studies: fine-mining the microbiome. Nature Reviews Microbiology 14, 508–522 (2016).
Gill, S. R. et al. Metagenomic Analysis of the Human Distal Gut Microbiome. Science 312, 1355–1359 (2006).
Heintz-Buschart, A. & Wilmes, P. Human Gut Microbiome: Function Matters. Trends in Microbiology 26, 563–574 (2018).
Young, V. B. The role of the microbiome in human health and disease: an introduction for clinicians. BMJ j831, https://doi.org/10.1136/bmj.j831 (2017).
Mason, M. R., Nagaraja, H. N., Camerlengo, T., Joshi, V. & Kumar, P. S. Deep Sequencing Identifies Ethnicity-Specific Bacterial Signatures in the Oral Microbiome. PLoS One 8, e77287 (2013).
Hall, A. B., Tolonen, A. C. & Xavier, R. J. Human genetic variation and the gut microbiome in disease. Nature Reviews Genetics 18, 690–699 (2017).
Yatsunenko, T. et al. Human gut microbiome viewed across age and geography. Nature 486, 222–227 (2012).
Odamaki, T. et al. Age-related changes in gut microbiota composition from newborn to centenarian: a cross-sectional study. BMC Microbiology 16 (2016).
Turnbaugh, P. J. et al. The Effect of Diet on the Human Gut Microbiome: A Metagenomic Analysis in Humanized Gnotobiotic Mice. Science Translational Medicine 1, 6ra14–6ra14 (2009).
David, L. A. et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563 (2013).
Tyakht, A. V. et al. Human gut microbiota community structures in urban and rural populations in Russia. Nature Communications 4 (2013).
Allendorf, K. Going Nuclear? Family Structure and Young Women’s Health in India, 1992–2006. Demography 50, 853–880 (2012).
Garceau, A., Wideroff, L., McNeel, T., Dunn, M. & Graubard, B. I. Population Estimates of Extended Family Structure and Size. Public Health Genomics 11, 331–342 (2008).
Kumbhare, S. V. et al. A cross-sectional comparative study of gut bacterial community of Indian and Finnish children. Scientific Reports 7 (2017).
Bhute, S. S. et al. Gut Microbial Diversity Assessment of Indian Type-2-Diabetics Reveals Alterations in Eubacteria, Archaea, and Eukaryotes. Frontiers in Microbiology 8 (2017).
Patil, D. P. et al. Molecular analysis of gut microbiota in obesity among Indian individuals. Journal of Biosciences 37, 647–657 (2012).
Bodkhe, R. et al. Comparison of Small Gut and Whole Gut Microbiota of First-Degree Relatives With Adult Celiac Disease Patients and Controls. Frontiers in Microbiology 10 (2019).
Chaudhari, D. et al. Understanding the association between the human gut, oral and skin microbiome and the Ayurvedic concept of prakriti. Journal of Biosciences 44 (2019).
Das, B. et al. Analysis of the Gut Microbiome of Rural and Urban Healthy Indians Living in Sea Level and High Altitude Areas. Scientific Reports 8 (2018).
Dehingia, M. et al. Gut bacterial diversity of the tribes of India and comparison with the worldwide data. Scientific Reports 5 (2015).
De Filippo, C. et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proceedings of the National Academy of Sciences 107, 14691–14696 (2010).
Bhute, S. et al. Molecular Characterization and Meta-Analysis of Gut Microbial Communities Illustrate Enrichment of Prevotella and Megasphaera in Indian Subjects. Frontiers in Microbiology 7 (2016).
Kulkarni, A. S., Kumbhare, S. V., Dhotre, D. P. & Shouche, Y. S. Mining the Core Gut Microbiome from a Sample Indian Population. Indian Journal of Microbiology 59, 90–95 (2018).
Chen, T. et al. Fiber-utilizing capacity varies in Prevotella- versus Bacteroides-dominated gut microbiota. Scientific Reports 7 (2017).
Gomez, A. et al. Host Genetic Control of the Oral Microbiome in Health and Disease. Cell Host & Microbe 22, 269–278.e3 (2017).
Gao, L. et al. Oral microbiomes: more and more importance in oral cavity and whole body. Protein & Cell 9, 488–500 (2018).
Takeshita, T. et al. Bacterial diversity in saliva and oral health-related conditions: the Hisayama Study. Scientific Reports 6 (2016).
Johansson, I., Witkowska, E., Kaveh, B., Lif Holgerson, P. & Tanner, A. C. R. The Microbiome in Populations with a Low and High Prevalence of Caries. Journal of Dental Research 95, 80–86 (2015).
Chen, H. & Jiang, W. Application of high-throughput sequencing in understanding human oral microbiome related with health and disease. Frontiers in Microbiology 5 (2014).
Zhao, H. et al. Variations in oral microbiota associated with oral cancer. Scientific Reports 7 (2017).
Oh, J., Byrd, A. L., Park, M., Kong, H. H. & Segre, J. A. Temporal Stability of the Human Skin Microbiome. Cell 165, 854–866 (2016).
Findley, K. & Grice, E. A. The Skin Microbiome: A Focus on Pathogens and Their Association with Skin Disease. PLoS Pathogens 10, e1004436 (2014).
Schommer, N. N. & Gallo, R. L. Structure and function of the human skin microbiome. Trends in Microbiology 21, 660–668 (2013).
Klymiuk, I., Bambach, I., Patra, V., Trajanoski, S. & Wolf, P. 16S Based Microbiome Analysis from Healthy Subjects’ Skin Swabs Stored for Different Storage Periods Reveal Phylum to Genus Level Changes. Frontiers in Microbiology 7 (2016).
Castelino, M. et al. Optimisation of methods for bacterial skin microbiome investigation: primer selection and comparison of the 454 versus MiSeq platform. BMC Microbiology 17 (2017).
Lozupone, C. A., Stombaugh, J. I., Gordon, J. I., Jansson, J. K. & Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 489, 220–230 (2012).
Koh, A., De Vadder, F., Kovatcheva-Datchary, P. & Bäckhed, F. From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell 165, 1332–1345 (2016).
Morotomi, M., Nagai, F., Sakon, H. & Tanaka, R. Dialister succinatiphilus sp. nov. and Barnesiella intestinihominis sp. nov., isolated from human faeces. International Journal of Systematic and Evolutionary Microbiology 58, 2716–2720 (2008).
Xie, H. et al. Intergeneric Communication in Dental Plaque Biofilms. Journal of Bacteriology 182, 7067–7069 (2000).
Kreth, J., Merritt, J. & Qi, F. Bacterial and Host Interactions of Oral Streptococci. DNA and Cell Biology 28, 397–403 (2009).
Singh, R. K. et al. Influence of diet on the gut microbiome and implications for human health. Journal of Translational Medicine 15 (2017).
Buford, T. W. (Dis)Trust your gut: the gut microbiome in age-related inflammation, health, and disease. Microbiome 5 (2017).
Nicoletti, C. Age-associated changes of the intestinal epithelial barrier: local and systemic implications. Expert Review of Gastroenterology & Hepatology 9, 1467–1469 (2015).
Man, A. L. et al. Age-associated modifications of intestinal permeability and innate immunity in human small intestine. Clinical Science 129, 515–527 (2015).
Tandon, D. et al. A snapshot of gut microbiota of an adult urban population from Western region of India. PLoS One 13, e0195643 (2018).
Louis, P., Hold, G. L. & Flint, H. J. The gut microbiota, bacterial metabolites and colorectal cancer. Nature Reviews Microbiology 12, 661–672 (2014).
Scott, K. P., Martin, J. C., Campbell, G., Mayer, C.-D. & Flint, H. J. Whole-Genome Transcription Profiling Reveals Genes Up-Regulated by Growth on Fucose in the Human Gut Bacterium “Roseburia inulinivorans”. Journal of Bacteriology 188, 4340–4349 (2006).
Shetty, S. A., Marathe, N. P., Lanjekar, V., Ranade, D. & Shouche, Y. S. Comparative Genome Analysis of Megasphaera sp. Reveals Niche Specialization and Its Potential Role in the Human Gut. PLoS One 8, e79353 (2013).
Dimitriu, P. A. et al. New Insights into the Intrinsic and Extrinsic Factors That Shape the Human Skin Microbiome. mBio 10 (2019).
Guinane, C. M. & Cotter, P. D. Role of the gut microbiota in health and chronic gastrointestinal disease: understanding a hidden metabolic organ. Therapeutic Advances in Gastroenterology 6, 295–308 (2013).
Rodríguez, J. M. et al. The composition of the gut microbiota throughout life, with an emphasis on early life. Microbial Ecology in Health & Disease 26 (2015).
Zhang, Y.-J. et al. Impacts of Gut Bacteria on Human Health and Diseases. International Journal of Molecular Sciences 16, 7493–7519 (2015).
Belkaid, Y. & Hand, T. W. Role of the Microbiota in Immunity and Inflammation. Cell 157, 121–141 (2014).
Afra, K., Laupland, K., Leal, J., Lloyd, T. & Gregson, D. Incidence, risk factors, and outcomes of Fusobacterium species bacteremia. BMC Infectious Diseases 13 (2013).
Woodmansey, E. J., McMurdo, M. E. T., Macfarlane, G. T. & Macfarlane, S. Comparison of Compositions and Metabolic Activities of Fecal Microbiotas in Young Adults and in Antibiotic-Treated and Non-Antibiotic-Treated Elderly Subjects. Applied and Environmental Microbiology 70, 6113–6122 (2004).
Singh, P. & Manning, S. D. Impact of age and sex on the composition and abundance of the intestinal microbiota in individuals with and without enteric infections. Annals of Epidemiology 26, 380–385 (2016).
Andreescu, C. et al. Age influence on periodontal tissues: a histological study. Romanian Journal of Morphology and Embryology 54, 811–815 (2013).
Wang, Y. & Kasper, L. H. The role of microbiome in central nervous system disorders. Brain, Behavior, and Immunity 38, 1–12 (2014).
Upadhyaya, S. & Banerjee, G. Type 2 diabetes and gut microbiome: at the intersection of known and unknown. Gut Microbes 6, 85–92 (2015).
Hartstra, A. V., Bouter, K. E. C., Bäckhed, F. & Nieuwdorp, M. Insights Into the Role of the Microbiome in Obesity and Type 2 Diabetes. Diabetes Care 38, 159–165 (2014).
Fadrosh, D. W. et al. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome 2, 6 (2014).
Magoc, T. & Salzberg, S. L. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963 (2011).
Zhernakova, A. et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science 352, 565–569 (2016).
Edgar, R. C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461 (2010).
McMurdie, P. J. & Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS One 8, e61217 (2013).
Taiyun, W. & Viliam, S. R package “corrplot”: Visualization of a Correlation Matrix (Version 0.84). Available at, https://github.com/taiyun/corrplot (2017).
Oksanen, J. Multivariate Analysis of Ecological Communities in R: Vegan Tutorial. Available at, http://cran.r-project.org (2013).
Lahti, L. et al. Tools for microbiome analysis in R. Version 1.5.23. 2017; Available at, http://microbiome.github.com/microbiome (2017).
Parks, D. H. & Beiko, R. G. Identifying biologically relevant differences between metagenomic communities. Bioinformatics 26, 715–721 (2010).
Heberle, H., Meirelles, G. V., da Silva, F. R., Telles, G. P. & Minghim, R. InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinformatics 16, 169 (2015).
Suryavanshi, M. V. et al. Hyperoxaluria leads to dysbiosis and drives selective enrichment of oxalate metabolizing bacterial species in recurrent kidney stone endures. Scientific Reports 6 (2016).
Acknowledgements
This work was supported by funding from the Department of Biotechnology, Government of India through a project entitled “PUNE MICROBIOME STUDY- Molecular analysis of human microbiome” (DBT Grant Number: BT/PR3461/BRB/10/968/2011). The authors would like to acknowledge the Director, KEMHRC, Pune and Director, NCCS, Pune for his support. The authors would like to thank Dr. Karen E. Nelson for her suggestions in the analysis and Dr. Padma Shastry and Abhijit Kulkarni for the English grammar corrections. Author D.S.C. would like to thank Shreyas Kumbhare and Sudarshan Shetty for their suggestions in the bioinformatics analysis, KEMHRC field staff for assisting in sample collection and the study subjects for their participation in the study.
Author information
Authors and Affiliations
Contributions
S.K.J., S.S., A.B., D. P. D. and Y.S.S. conceived the study. D.S.C., D.P.D., D.M.A., S.S., A.B., S.K.J. and Y.S.S. designed the study. D.M.A., D.B., P.J., A.H.G. and S.K.J. provided the samples. D.S.C. performed the experiments. A.H.G., D.B. and P.J assisted in the experiments. Y.S.S., S.J., V.P.S. and U.K.P. provided guidance with the experiments. D.S.C. and D.P.D. performed the bioinformatics and statistical analyses. D.M. helped in that data analysis. H.L. guided in analysis of routine diet of the study subjects. D.S.C. and D.P.D. drafted the manuscript with input from V.P.S., Y.S.S. and S.K.J. All authors contributed to manuscript revisions, have read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chaudhari, D.S., Dhotre, D.P., Agarwal, D.M. et al. Gut, oral and skin microbiome of Indian patrilineal families reveal perceptible association with age. Sci Rep 10, 5685 (2020). https://doi.org/10.1038/s41598-020-62195-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-62195-5
This article is cited by
-
Gut microbial ecology and exposome of a healthy Pakistani cohort
Gut Pathogens (2024)
-
The oral microbiome, pancreatic cancer and human diversity in the age of precision medicine
Microbiome (2022)
-
Altered Salivary Microbiota Following Bifidobacterium animalis Subsp. Lactis BL-11 Supplementation Are Associated with Anthropometric Growth and Social Behavior Severity in Individuals with Prader-Willi Syndrome
Probiotics and Antimicrobial Proteins (2022)
-
Taxonomic profiling and functional characterization of the healthy human oral bacterial microbiome from the north Indian urban sub-population
Archives of Microbiology (2021)
-
Comparative analysis of human facial skin microbiome between topical sites compared to entire face
Genes & Genomics (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.