Abstract
With the development of data mining, machine learning offers opportunities to improve discrimination by analyzing complex interactions among massive variables. To test the ability of machine learning algorithms for predicting risk of type 2 diabetes mellitus (T2DM) in a rural Chinese population, we focus on a total of 36,652 eligible participants from the Henan Rural Cohort Study. Risk assessment models for T2DM were developed using six machine learning algorithms, including logistic regression (LR), classification and regression tree (CART), artificial neural networks (ANN), support vector machine (SVM), random forest (RF) and gradient boosting machine (GBM). The model performance was measured in an area under the receiver operating characteristic curve, sensitivity, specificity, positive predictive value, negative predictive value and area under precision recall curve. The importance of variables was identified based on each classifier and the shapley additive explanations approach. Using all available variables, all models for predicting risk of T2DM demonstrated strong predictive performance, with AUCs ranging between 0.811 and 0.872 using laboratory data and from 0.767 to 0.817 without laboratory data. Among them, the GBM model performed best (AUC: 0.872 with laboratory data and 0.817 without laboratory data). Performance of models plateaued when introduced 30 variables to each model except CART model. Among the top-10 variables across all methods were sweet flavor, urine glucose, age, heart rate, creatinine, waist circumference, uric acid, pulse pressure, insulin, and hypertension. New important risk factors (urinary indicators, sweet flavor) were not found in previous risk prediction methods, but determined by machine learning in our study. Through the results, machine learning methods showed competence in predicting risk of T2DM, leading to greater insights on disease risk factors with no priori assumption of causality.
Similar content being viewed by others
Introduction
Type 2 diabetes mellitus (T2DM) is a long-term metabolic disorder with high morbidity in humans around the world. The prevalence of diabetes is increasing rapidly worldwide, including in China1. In China, diabetes was estimated to affect 144.4 million people aged 20–79 according to the report of the international diabetes federation in 20172. The prevalence of diabetes in a rural population of Henan province is high which can be seen in the Henan Rural Cohort Study3. Although diabetes is an irreversible disease, it is largely preventable4. The risk of developing diabetes will be reduced through early detection and lifestyle interventions. For individual patient care, physicians are well prepared to identify those at risk for T2DM. However, when trying to screen thousands of patients with high-risk conditions, the challenges faced by physicians become apparent. There is a need for analytics techniques to assist in T2DM mass screening.
Many risk scores based on statistical knowledge have been developed for predicting individual’s risk of developing T2DM, such as risk evaluation formula5, Archimedes trial-validated diabetes model6, the diabetes risk score7, genetic risk score8, the New Chinese Diabetes Risk Score42 and the American Academy of Family Physicians risk model9. These methods made the implicit assumption that each risk factor was linear to the outcome. The complex relationships between nonlinear interaction factors might be oversimplified, leading to the potential loss of related information10,11. Moreover, when the number of variables increased, the hypothesis testing method became complicated12. In contrast to traditional methods, machine learning can learn the nonlinear interactions iteratively from large amounts of data using computer algorithms13, which have been applied in various fields, such as disease risk assessment and prediction14,15. Recent research shows that machine learning methods can describe patients’ characteristics and identify patients at risk of developing T2DM16,17. A study illustrated the performance of support vector machine for detecting persons with diabetes and pre-diabetes18. To assess the ability to estimate the risk of developing T2DM, a study evaluated the performance of different machine learning and statistical techniques, and the experimental results showed the comprehensive performance the ensembles of ANN was better than other models19. A data mining pipeline based on classification algorithm was built to predict T2DM complications based on electronic health record data from nearly one thousand patients, which showed the validity of machine learning method20. An ensemble approach with the use of the vote method with three Decision Trees was developed to predict incident diabetes using 13 attributes21, and improved the value of AUC to 0.922. A novel joint clustering and classification (JCC) method which could discover hidden clusters features in the patient samples was developed to predict diabetes, and the method performed best among the methods that were applicable to the interpretation of prediction22. A study used neural network, decision tree, and random forest to predict diabetes mellitus with 14 attributes, and the results showed that the highest accuracy method was random forest23. Another study compared the performance of several machine learning techniques to predict the risk of developing T2DM in short, medium, and long term, and the results showed that logistic regression outperformed in short, medium term while support vector machines presented better performance in long term24. A machine learning-based framework for identifying subjects with T2DM from EHR was constructed via feature engineering, and the results revealed that the framework performed higher identification compared with the expert algorithm25.
However, the current methods just focused on performance comparison of prediction techniques with fixed number of variables, and they were also done on a small population sample. To date, there has been no large-scale investigation applying machine-learning for risk assessment in the general rural population. Therefore, the purpose of this study was to (1) evaluate an array of machine learning algorithms for predicting the risk of T2DM in a rural Chinese population; (2) identify the important variables, and (3) reveal the model performance of each model on a varying number of variables.
Method
Study participants
The participants of this study came from the Henan Rural Cohort Study (Registration number: ChiCTR-OOC-15006699). A total of 39259 participants aged between 18 to 79 years were recruited from five rural areas in Henan province of China over the period between July 2015 and September 2017. The design and population characteristics of the study have been described in the previous articles26,27,28. Data on socio-demographic characteristics, information on physical examination, and laboratory test data were collected. Participants were excluded if they: (1) were diagnosed with kidney failure (N = 18) or cancer (N = 332); (2) had type 1 diabetes mellitus (N = 4); (3) had gestational diabetes mellitus (N = 634); (4) had incomplete information on diagnoses of T2DM (N = 63); and (5) had incomplete information of potential covariates (n = 2127). Finally, 36,652 participants were included for the present study.
Definition of T2DM
After excluding participants with type 1 diabetes mellitus, gestational diabetes mellitus, and other special type diabetes, T2DM was a self-reported previous diagnosis of diabetes by a physician or fasting plasma glucose level ≥7.0 mmol/L according to the American Diabetes Association (ADA) diagnostic criteria29.
Machine learning methods
We used logistic regression, artificial neural networks, classification and regression tree, support vector machine, and ensemble learning (random forest and gradient boosting machine) to build the risk assessment model. From the description of basic characteristics of non-T2DMs and T2DMs, the data is imbalanced. The model is likely to be biased towards the dominant class, with poor accuracy in classifying negative cases. In view of this problem, the Synthetic Minority Over-Sampling Technique (SMOTE)24,30,31 algorithm was used to address the data. All models were constructed using the package sklearn (0.21.3) of Python 3.7 programing language.
Artificial neural networks
Artificial neural networks32 are computing systems that are based on the neurons of the human brain. ANN can learn all complex and non-linear interactions between variables to look for patterns in the data. ANN is divided into multi-hidden layer neural network and single hidden layer neural network. Each layer contains a number of neurons connected by directed arcs with variable weights. In our study, the neural network consists of three layers: an input layer to accept all risk factors, a hidden layer to process information and an output layer to calculate responses.
Classification and regression tree
A decision tree is a tree structure in which each internal node represents a test on an attribute, each branch represents a test output, and each leaf node represents a category33. Typical algorithms of decision tree include ID3, C4.5, CART, and so on. Considering the extensive application of CART in clinical and basic research, we used CART in this study34. CART is a non-parametric decision tree learning technology, which generates a classification tree or regression tree according to whether the dependent variable is classified or numerical35.
Logistic regression
Logistic regression (LR) is a generalized linear regression analysis model, which works to find the best fitting model that can describe the relationship between dependent variables and independent predictors36. LR model is most widely used when people are interested in predicting disease or health status37. The LR model can compute the probability of an individual developing T2DM based on the risk factors input. If a subject suffers from T2DM, the value of Y is 1; otherwise, Y is 0. We defined the probability of an individual developing T2DM is \(\,p(Y=1|X)=p(X)\). Then, the formula of the LR model is defined as follows.
and equivalently, after exponentiating both sides:
The probability of an individual developing T2DM is
Where \(X=({X}_{1},{X}_{2}\cdots {X}_{k})\) represents the risk factors, \(\beta =({\beta }_{1},{\beta }_{2}\cdots {\beta }_{k})\) are the coefficients estimated by using the method of maximum likelihood.
Support vector machine
Support vector machine (SVM) is a kind of generalized linear classifier that classifies binary data according to supervised learning. Its decision boundary is the maximum margin hyper plane for the positive and negative classes38. In our study, each data sample is made of 60 features. The value of each feature is a vector of a particular dimension. Then, we used SVM to construct a hyperplane in a high-dimensional space, which can distinguish the two classes nicely.
Ensemble learning
Ensemble learning is an algorithm that combines basic learners such as decision trees and linear classifiers. The main idea of ensemble learning is to use multiple learning algorithms to achieve better performance than any constituent learning algorithm alone. Common types of ensembles are boosting, bagging, random subspace.
Random forest (RF) is an algorithm combines bagging ensemble learning theory with random subspace approach. RF generates many decision trees for splitting data randomly at training time. For each node of the base decision tree, a subset containing K attributes is randomly selected from the attribute set of that node, and then an optimal attribute is selected from the sub-set for partitioning. Each tree provides a classification as a vote for each tree, and the RF ultimately chooses the classification with the most votes39.
Gradient boosting machine (GBM) is an iterative algorithm whose core idea is to train different classifiers (weak classifiers) for the same training set, and then combine these weak classifiers to form a stronger final classifier (strong classifier). Through a series of iterations to optimize the classification results, each iteration is introduced into a weak classifier, to overcome the existing shortcomings of weak classifier combination. GBM is based on the residual of training data fitted by the previous weak classifier to enhance the model when training each weak classifier. Compared with most learning algorithms, it is less prone to over fitting.
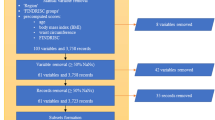
Figure 1 showed the methodology of this study. In this study, risk assessment models for T2DM were developed using 6 ML algorithms on all variables. Next, algorithms were iteratively introduced to a growing number of ranked variables (5/10/15/…) selected by the algorithm itself. All models were trained and tested by 10-fold cross-validation during each iteration process, which was repeated 100 times. Performance of all models was calculated on the test samples. All models’ parameters were determined using 10-fold cross-validation and grid search on the training data (Supplementary Table 2).
Methodology. Abbreviation: LR, logistic regression; CART, classification and regression tree; GBM, gradient boosting machine; ANN, artificial neural network; RF, Random forest; SVM, Support vector machine; PPV, positive predictive value; NPV, negative predictive value; AUC, area under curve; AUPR, area under precision recall curve.
Statistical analysis and evaluation on the model
Model performance: Discrimination refers to the model’s ability to identify who is at risk of developing T2D and who is not. We used sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), area under precision recall curve (AUPR) and area under curve (AUC) to evaluate discrimination. Sensitivity is a synonym for recall rate, true positive rate, and represents the proportion of real positive samples that are identified correctly. For instance, in our study, the subject who diagnosed with T2DM was defined as 1, namely the positive sample. Otherwise, it was a negative sample (0). Specificity indicates the rate of real negative samples can be detected correctly. PPV stands for the proportion of positive results in diagnostic tests that are true positive results. NPV is the proportion of negatives in diagnostic tests that are true negative results. For binary classification models, AUC and AUPR were also used to evaluate the performance.
Variable importance: For the diabetes study, we also listed the importance of variables. For LR and SVM models, variable importance was determined by the coefficient effect size. CART model estimated the variable importance by summing changes in the mean squared error due to splits on every variable and dividing the sum by the number of branch nodes. Variable importance of RF was estimated by permutation of out-of-bag variable observations. GBM computed the variable importance by summing these estimates over all weak learners in the classification ensemble method. ANN used overall connection weights of variable to filter variables40. In order to combine the variable importance of each method, the variable importance was also estimated using shapley additive explanations approach, which is a unified approach to explain the output of any machine learning model41.
Categorical variables were described as percentages (%), and continuous variables were shown as mean ± standard deviation (SD). Differences in the characteristics of T2DM and Non-T2DM groups were determined with the chi-square test for categorical variables and t-test for continuous variables. All statistical analyses were performed using SPSS (v.21, IBM) and a two-tailed P value < 0.05 was considered statistically significant.
Ethics approval
Ethics approval was obtained from the “Zhengzhou University Life Science Ethics Committee”, and written informed consent was obtained for all participants. Ethic approval code: [2015] MEC (S128). The present study was conducted in accordance with the guidelines of the Declaration of Helsinki.
Results
Basic characteristics
The general characteristics of the study population were presented in Table 1. The study population consisted of 14,375 men and 22,277 women. Compared with participants without T2DM, individuals with T2DM tended to be with higher age, BMI, waist circumference, heart rate, waist to height ratio, urine glucose, low-density lipoprotein cholesterol, and were more likely to have a family history of T2DM, hypertension, coronary heart disease. In contrast, among participants without T2DM, higher creatinine, higher high-density lipoprotein cholesterol, and higher uric acid were more common. Further details were presented in Supplementary Table 1.
Variable importance analysis
The top 10 variables according to the variable importance of each algorithm were presented in Table 2 (Supplementary Table 3). Elevated urine glucose level was presented as top-ranked variables by all algorithms. Indicators of obesity repeatedly were appeared at the top of the list, such as waist to hip ratio, and waist to height ratio. This phenomenon confirmed that obesity is a risk factor of T2DM. Hypertension was ranked as an important factor of T2DM by most models, perhaps reflecting the relationship between hypertension and the development of T2DM. The risk factors in the New Chinese Diabetes Risk Score included sex, age, family history of diabetes, waist circumference, BMI, SBP. Several risk factors of the New Chinese Diabetes Risk Score (age, family history of diabetes, sex, and SBP) were shown in the list of top-ranked variables in our study. Common variables for diabetes were also identified by machine learning methods, such as genetic factors, hypertension, insulin, and so on. Also, new important variables (urinary parameters) were not found in previous risk prediction methods but determined by machine learning. Furthermore, the LR, SVM and ANN models prioritized genetic factor and urinary parameters, such as T2DM history of mother/father, urine glucose, urine protein, and so on.
We analyzed the importance of variables based on all models using the shapley additive explanations approach (Supplementary Table 4). As shown in Table 3. Among the top-10 variables across all methods were sweet flavor, urine glucose, age, heart rate, creatinine, waist circumference, uric acid, pulse pressure, insulin, and hypertension.
Comparison of model performance
Table 4 presented the comparison results of machine learning algorithms. Using all available variables, all models for predicting risk of T2DM demonstrated strong predictive performance, with AUCs ranging between 0.811 and 0.872. The GBM model performed best (AUC = 0.872 with laboratory variables), and also presented better specificity (81.71%), positive predictive value (28.83%), and AUPR (0.546). In terms of accuracy and negative predictive value, the data showed that RF model remained strong predictive performance (85.90%, and 97.52% respectively). ANN model’s sensitivity was best among all models’. Using only non-laboratory data, such as BMI, age resulted in large declines in model performance. Moreover, the models with only non-laboratory data were also significantly better than the New Chinese Diabetes Risk Score42 based on statistical knowledge only using non-laboratory data (AUC = 0.728, p < 0.05) (Supplementary Figure 1).
Figure 2 displayed the receiver operator characteristic curves of each model with all variables. This visualization revealed that the GBM performed similarly to RF model, and the two models exhibited greater superiority than the ANN model (p < 0.05), with 0.872, 0.868 and 0.858 respectively. The above three models (GBM, RF and ANN) performed significantly better than CART (AUC = 0.11), LR (AUC = 0.841), and SVM (AUC = 0.835) (p < 0.05). The area under precision recall curve also showed the same results (Figure 3).
Model performance with a varying number of variables
In order to compare the performance of different models on a varying number of variables, the highest 5/10/15… ranked variables of each model were consecutively incorporated into each model. As shown in Figure 4. Overall, as the number of variables increased, the graph showed the increase of AUC values except the CART model. Before introducing with 30 variables, the LR, GBM, ANN, SVM, and RF models presented dramatic rise trends on the values of AUC. Performance of the five models plateaued when introduced 30 variables to each model. After that, all trends showed slight fluctuation, but the changes were modest. The CART model maintained a constant trend of AUC value.
Discussion
Using machine learning methods, this study developed several risk assessment models for characterizing the risk of developing T2DM. High predictive performance was achieved by all models, with AUCs ranging from 0.811to 0.872. Compared to other models, the GBM model performed the best, with an AUC value of 0.872 (95% 0.858–0.886) and the models’ performance significantly better than the traditional risk score. In addition to common factors for diabetes, new important factors (urinary parameters) were not found in previous risk assessment methods, but determined by machine learning in our study. Our study demonstrated that machine learning technologies are uniquely positioned to identify significant risk factors in large-scale epidemiological studies.
To our knowledge, this is the first study to assess the importance of variables and characterize the risk of developing T2DM with use of different machine learning methods in a Chinese rural population. Our results were consistent with the previous findings. The New Chinese Diabetes Risk Score showed that sex, age, family history of diabetes, waist circumference, BMI, SBP were important risk factors42. Our results also revealed their prominent presence on the top-10 key factors for T2DM. Our data also indicated that obesity was a major risk factor for the development of T2DM43. The previous studies have demonstrated the significant role of boosting method in other medical fields, such as urinary tract infections44, hepatocellular carcinoma diagnosis45, prediction of hip fracture46. Our results confirmed the outstanding performance of the boosting method in the risk assessment of T2DM.
Identifying the key factors is of great clinical significance in the risk assessment of T2DM. The severity of T2DM is often estimated through a lot of factors in different aspects, including socio-demographic characteristics, anthropometric measures and laboratory test data. Given that the diversity and massive of factors in the development phase of T2DM, it is difficult to choose a specific number of variables for risk assessment. Compared to models with no laboratory data, the inclusion of laboratory data resulted in significant increase in the identification capabilities of models. This phenomenon shows that adding effective laboratory data can help identify the risk of T2DM patients. Our study also showed that the importance of different factors depended on the modeling technique. For LR, SVM and ANN models, the genetic factors and urinary indicators, such as diabetes history of mother/father, urine glucose, occupied center stage in the risk assessment of T2DM. Moreover, our results showed 30–35 variables were needed when the model performance reached a plateau, and the model performance would not be improved with too many variables. Over the past decade, the ability to collect data has become faster and cheaper, but we need to pay more attention to the model with too many features.
Data analysis techniques or model fitting ability are important in disease risk assessment and prediction. With the use of traditional statistical approaches, many risk scores and prediction models have been developed based on logistic regression. If the relationship among the data is linearly separable, traditional methods will fit better47,48. If not, such models may oversimplify complex relationships among factors with nonlinear interactions, leading to the potential loss of important relevant information. This suggests the important of choosing an appropriate model according to the characteristics of the data set. According to the data characteristics of the Henan Rural Cohort Study, our results showed the boosting model fit data best.
The performance of diagnosis model based on machine learning will be better if the number of training samples is large49. Compared to previous studies, the major strength of our study was the relatively large sample size including 36652 subjects from the rural population in China. Also, we compared the model performance from two aspects: the fixed number of variables and the dynamic number of variables, which confirmed models with several variables could perform no worse than the model with all variables50. Furthermore, the superiority and feasibility of nonparametric algorithms were proved compared with the model based on logistic regression.
However, several limitations should be worth mentioning. Firstly, the research findings were derived from a cross-sectional study without follow-up data; therefore, we may not be able to determine the causal and temporal associations. Secondly, we need to do future research with external validation and other machine learning methods to assess the model performance. In addition, it’s difficult to explain the inherent complexity of variable interactions and their impacts on outcomes due to the “black box” nature of machine learning methods.
In conclusion, using a series of machine learning models, we developed a data mining approach to characterize risk ofT2DM and compared the model performance from the fixed number of variables and the dynamic number of variables. Our results showed the advantage ability of machine learning to identify risk factors and predict outcomes across a wide range of data and an increasing number of variables, which leading to greater insights on disease risk factors with no prior assumption of causality.
Data sharing statement
All relevant data are within the paper and its Supporting Information files. Contact to Dr. Chongjian Wang (tjwcj2005@126.com) for additional information regarding data access.
References
Xu, Y. et al. Prevalence and control of diabetes in Chinese adults. JAMA. 310, 948–59 (2013).
International Diabetes Federation. IDF diabetes atlas. 8th. http://www.diabetesatlas.org/ (2017).
Liu, X. et al. Prevalence, awareness, treatment, control of type 2 diabetes mellitus and risk factors in Chinese rural population: the RuralDiab study. Sci. Rep. 6, 31426 (2016).
Li, Y. et al. Time trends of dietary and lifestyle factors and their potential impact on diabetes burden in china. Diabetes Care. 40, 1685–1694 (2017).
Collins, G. S., Mallett, S., Omar, O. & Yu, L. Developing risk prediction models for type 2 diabetes: a systematic review of methodology and reporting. BMC Med. 9, 1–14 (2011).
Eddy, D. M. & Schlessinger, L. Archimedes: a trial-validated model of diabetes. Diabetes Care. 26, 3093–3101 (2003).
Lindstrom, J. & Tuomilehto, J. The Diabetes Risk Score: a practical tool to predict type 2 diabetes risk. Diabetes Care. 26, 725–731 (2003).
Cornelis, M. C. et al. Joint effects of common genetic variants on the risk for type 2 diabetes in U.S. men and women of European ancestry. Ann. Intern. Med. 150, 541–550 (2009).
Pippiti, K., Li, M. & Gurgle, H. Diabetes mellitus: screening and diagnosis. Am. Fam. Phys. 93, 103–9 (2016).
Obermeyer, Z. & Emanuel, E. J. Predicting the Future—Big Data, Machine Learning, and Clinical Medicine. N. Engl. J. Med. 375, 1216–9 (2016).
Harrell, F. Regression Modeling Strategies: With Applications to Linear Models, Logistic and Ordinal Regression, and Survival Analysis, Springer. (2015).
Dag, A., Oztekin, A., Yucel, A., Bulur, S. & Megahed, F. M. Predicting heart transplantation outcomes through data analytics. Decis. Support Syst. 94, 42–52 (2017).
Lagani, V., Koumakis, L., Chiarugi, F., Lakasing, E. & Tsamardinos, I. A systematic review of predictive risk models for diabetes complications based on large scale clinical studies. J. Diabetes Complications 27, 407–413 (2013).
Deo, R. C. Machine learning in medicine. Circulation 132, 1920–1930 (2015).
Ambale-Venkatesh, B. et al. Cardiovascular event prediction by machine learning: The Multi-Ethnic Study of Atherosclerosis. Circ. Res. 121, 1092–1101 (2017).
Dinh, A., Miertschin, S., Young, A. & Mohanty, S. D. A data-driven approach to predicting diabetes and cardiovascular disease with machine learning. BMC Med. Inform. Decis. Mak. 19, 211 (2019).
Ramezankhani, A. et al. Applying decision tree for identification of a low risk population for type 2 diabetes. Tehran Lipid and Glucose Study. Diabetes research and clinical practice 105, 391–398 (2014).
Yu, W., Liu, T., Valdez, R., Gwinn, M. & Khoury, M. J. Application of support vector machine modeling for prediction of common diseases: the case of diabetes and pre-diabetes. BMC Med. Inform. Decis. Mak. 10, 16 (2010).
Dalakleidi, K., Zarkogianni, K., Thanopoulou, A. & Nikita, K. Comparative assessment of statistical and machine learning techniques towards estimating the risk of developing type2 diabetes and cardiovascular complications. Expert Systems. 34, e12214 (2017).
Dagliati, A. et al. Machine learning methods to predict diabetes complications. J. Diabetes Sci. Technol. 12, 295–302 (2018).
Alghamdi, M. et al. Predicting diabetes mellitus using SMOTE and ensemble machine learning approach: the Henry Ford ExercIse Testing (FIT) project. PLoS ONE. 12, e0179805 (2017).
Brisimi, T. S. et al. Predicting Chronic Disease Hospitalizations from Electronic Health Records: An Interpretable Classification Approach. Proceedings of the IEEE 106, 690–707 (2018).
Zou, Q. et al. Predicting Diabetes Mellitus With Machine Learning Techniques. Front. Genet. 9, 515 (2018).
Amir, T. K. & Wilson, J. M. Identifying People at Risk of Developing Type 2 Diabetes: A Comparison of Predictive Analytics Techniques and Predictor Variables. International Journal of Medical Informatics. 119, 22–38 (2018).
Zheng, T. et al. A machine learning-based framework to identify type 2 diabetes through electronic health records. International Journal of Medical Informatics. 7, 120–127 (2017).
Liu, X. et al. The Dynamics of Type 2 Diabetes Mellitus Prevalence and Management Rates among Rural Population in Henan Province, China. Journal of Diabetes Research 2017, 1–9 (2017).
Li, N. et al. Associations of long-term exposure to ambient PM1 with hypertension and blood pressure in rural Chinese population: The Henan rural cohort study. Environ Int. 128, 95–102 (2019).
Liu, X. et al. The Henan Rural Cohort: a prospective study of chronic non-communicable diseases. Int J Epidemiol. 48, 1756–1756j (2019).
American Diabetes Association. Diagnosis and classifcation of diabetes mellitus. Diabetes care. 32, Suppl 1, S62–S67 (2009).
Chawla, N. V., Bowyer, K. W., Hall, L. O. & Kegelmeyer, W. P. SMOTE: synthetic minority over-sampling technique. J. Artif. Intell. Res. 16, 321–57 (2002).
Chawla, N. V. Data mining for imbalanced datasets: an overview. Data Mining and Knowledge Discovery Handbook, 853–867(2005).
Amato, F. et al. Artificial neural networks in medical diagnosis. Journal of Applied Biomedicine. 11, 47–58 (2013).
Liao, Z. J., Wan, S., He, Y. & Zou, Q. Classification of small GTPases with hybrid protein features and advanced machine learning techniques. Curr.Bioinform. 13, 492–500 (2018).
Trendowicz A., Jeffery R. Classification and Regression Trees. In: Software Project Effort Estimation. Springer, Cham. 295–304 (2014).
Esmaily, H. et al. A Comparison between Decision Tree and Random Forest in Determining the Risk Factors Associated with Type 2. Diabetes. J. Res. Health Sci. 18, e00412 (2018).
Bagley, S. C., White, H. & Golomb, B. A. Logistic regression in the medical literature: standards for use and reporting, with particular attention to one medical domain. J. Clin. Epidemiol. 54, 979–985 (2001).
Kengne, A. P. et al. Non-invasive risk scores for prediction of type 2 diabetes (EPIC-InterAct): a validation of existing models. The Lancet Diabetes & Endocrinology 2, 19–29 (2014).
Noble, W. S. What is a support vector machine? Nature Biotechnology 24, 1565–1567 (2006).
Svetnik, V. et al. Random forest: a classification and regression tool for compound classification and QSAR modeling. J. Chem. Inform. Comput. Sci. 43, 1947–1958 (2003).
Weng, S. F., Reps, J., Kai, J., Garibaldi, J. M. & Qureshi, N. Can machine-learning improve cardiovascular risk prediction using routine clinical data? PLoS ONE 12, e0174944 (2017).
Lundberg, S., Lee, S. I. A unified approach to interpreting model predictions. In Advances in Neural Information Processing Systems, 4765–4774 (2017).
Zhou, X. et al. Nonlaboratory-based risk assessment algorithm for undiagnosed type 2 diabetes developed on a nation-wide diabetes survey. Diabetes Care. 36, 3944–3952 (2013).
Tian, Z. et al. Gender-specific associations of body mass index and waist circumference with type 2 diabetes mellitus in Chinese rural adults: The Henan Rural Cohort Study. J Diabetes Complications. 32, 824–829 (2018).
Taylor, R. A., Moore, C. L., Cheung, K.-H. & Brandt, C. Predicting urinary tract infections in the emergency department with machine learning. PloS ONE. 13, e0194085 (2018).
Sato, M. et al. Machine-learning Approach for the Development of a Novel predictive Model for the Diagnosis of Hepatocellular Carcinoma. Sci. Rep. 9, 7704 (2019).
Kruse, C., Eiken, P. & Vestergaard, P. Machine Learning Principles Can Improve Hip Fracture Prediction. Calcified tissue international 100, 348–360 (2017).
Wu, X., Zhu, X., Wu, G. Q. & Ding, W. Data mining with big data. IEEE transactions on knowledge and data engineering 26, 97–107 (2014).
Hengl, S., Kreutz, C., Timmer, J. & Maiwald, T. Data-based identifiability analysis of non-linear dynamical models. Bioinformatics 23, 2612–2618 (2007).
Zacksenhouse, M., Braun, S., Feldman, M. & Sidahmed, M. Toward helicopter gearbox diagnostics from a small number of examples. Mechanical Systems and Signal Processing. 14, 523–543 (2000).
Yun, Y. H., Deng, B. C., Cao, D. S., Wang, W. T. & Liang, Y. Z. Variable importance analysis based on rank aggregation with applications in metabolomics for biomarker discovery. Analytica Chimica Acta 911, 27–34 (2016).
Acknowledgements
The authors thank all of the participants, coordinators, and administrators for their support and help during the research. This research was supported by the National Key Research and Development Program Precision Medicine Initiative of China (Grant NO: 2016YFC0900803), National Natural Science Foundation of China (Grant NO: 81573243, 81602925, 21806146), Henan Natural Science Foundation of China (Grant NO: 182300410293), Science and Technology Foundation for Innovation Talent of Henan Province (Grant NO: 164100510021), Science and Technology Innovation Talents Support Plan of Henan Province Colleges and Universities (Grant NO: 14HASTIT035). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
Z.F.W. and C.J.W. conceived and designed the study. L.Y.Z., M.M.N. and Y.K.W. coordinated data collection. L.Y.Z. and Y.K.W. conducted the analyses. L.Y.Z. wrote the manuscript. All co-authors critically revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhang, L., Wang, Y., Niu, M. et al. Machine learning for characterizing risk of type 2 diabetes mellitus in a rural Chinese population: the Henan Rural Cohort Study. Sci Rep 10, 4406 (2020). https://doi.org/10.1038/s41598-020-61123-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-61123-x
This article is cited by
-
Machine learning reduces soft costs for residential solar photovoltaics
Scientific Reports (2023)
-
An efficient transfer learning approach for prediction and classification of SARS – COVID -19
Multimedia Tools and Applications (2023)
-
Operationalising ethics in artificial intelligence for healthcare: a framework for AI developers
AI and Ethics (2023)
-
Design of optimal bidirectional long short term memory based predictive analysis and severity estimation model for diabetes mellitus
International Journal of Information Technology (2023)
-
Methodological guidelines to estimate population-based health indicators using linked data and/or machine learning techniques
Archives of Public Health (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.