Abstract
Many Gram-negative bacterial pathogens interact with mammalian cells by using type III secretion systems (T3SS) to inject virulence proteins into host cells. A subset of these injected protein ‘effectors’ are enzymes that inhibit the function of host proteins by catalyzing the addition of unusual post-translational modifications. The E. coli and Citrobacter rodentium NleB effectors, as well as the Salmonella enterica SseK effectors are glycosyltransferases that modify host protein substrates with N-acetyl glucosamine (GlcNAc) on arginine residues. This post-translational modification disrupts the normal functioning of host immune response proteins. T3SS effectors are thought to be inactive within the bacterium and fold into their active conformations after they are injected, due to the activity of chaperones that keep the effectors in a structural state permissive for secretion. While performing mass spectrometry experiments to identify glycosylation substrates of NleB orthologs, we unexpectedly observed that the bacterial glutathione synthetase (GshB) is glycosylated by NleB on arginine residue R256. NleB-mediated glycosylation of GshB resulted in enhanced GshB activity, leading to an increase in glutathione production, and promoted C. rodentium survival in oxidative stress conditions. These data represent, to our knowledge, the first intra-bacterial activity for a T3SS effector and show that arginine-GlcNAcylation, once thought to be restricted to host cell compartments, also plays an important role in regulating bacterial physiology.
Similar content being viewed by others
Introduction
The NleB (Escherichia coli and Citrobacter rodentium) and SseK (Salmonella enterica) enzymes are type three secretion system (T3SS) effector proteins that glycosylate host proteins with N-acetyl glucosamine (GlcNAc) on arginine residues to subvert their function in the innate immune system1,2,3. Arginine glycosylation is unusual because it occurs on the guanidinium groups of arginines, which are poor nucleophiles. The NleB/SseK orthologs share high degrees of structural similarity and consist of a catalytic domain including essential DXD and HEN motifs, a helix-loop-helix (HLH) domain, and a C-terminal lid domain4,5,6. Several ‘death domain’-containing proteins such as the Fas-Associated protein with Death Domain (FADD), tumor necrosis factor receptor type 1-associated death domain protein (TRADD), and the receptor interaction serine/threonine-protein kinase 1 (RIPK1) are NleB/SseK substrates2. These effectors disrupt tumor necrosis factor receptor (TNFR)-associated factor (TRAF) signaling, leading to inhibition of the pro-inflammatory NF-κB pathway1,2,3.
T3SS effectors are chaperoned in the bacterium after their synthesis to keep them partially unfolded and competent for secretion, as well as for targeting the effectors to the T3SS sorting platform7. The chaperones are then stripped from their effector substrates at the sorting platform and the effectors are secreted in an unfolded conformation8. T3SS effectors are generally believed to be inactive until they are injected into host cells, where they then fold into their active conformations9.
Some precedent for type IV secretion system (T4SS) effector activity in both the bacterium and the host may exist in plant pathogens. Agrobacterium tumefaciens transfers a nucleoprotein complex into plant cells. The VirD2 protein is associated with the transferred DNA (T-DNA). VirD2 has endonuclease activity within the bacterium to initiate T-DNA transfer10. VirD2 also targets the nucleoprotein complex in the plant cell nucleus, where it assists in integrating T-DNA into plant chromosomes11. Therefore, VirD2 may have enzymatic functions both within the bacterium and in the host plant cell. A recent study also indicated that Yersinia pseudotuberculosis uses the secreted T3SS translocator YopD to control RNA regulators and increase the abundance of LcrF, a common transcriptional activator of other T3SS effector genes12.
N-linked protein glycosylation on arginine has also been reported for the EarP glycosyltransferase from E. coli and Pseudomonas aeruginosa13. In this case, a single arginine rhamnosylation event activates the function of the polyproline-specific bacterial translation elongation factor EF-P13. Although the relatively inert guanidine group of arginine was previously thought to impede nucleophilic attack onto donor substrates, it is now clear that many bacterial enzymes have overcome this barrier. The enzymology and functional roles of arginine glycosylation, methylation, phosphorylation, and ADP-ribosylation were recently reviewed14.
While characterizing additional NleB glycosylation substrates, we made the unexpected observation that the bacterial glutathione synthetase (GshB) is glycosylated by NleB on an arginine residue. This glycosylation contributes to bacterial survival in hydrogen peroxide stress conditions by enhancing GshB activity and increasing intracellular levels of glutathione (GSH). Thus, NleB is active and performs important biological functions within the bacterium, prior to its secretion.
Results
NleB glycosylates GshB
We performed mass spectrometry experiments to identify new glycosylation substrates of NleB orthologs from EHEC strains associated with human disease outbreaks. These experiments were conducted by infecting HEK293T cells with EHEC strains that express NleB1. We then used an anti-Arg-R-GlcNAc monoclonal antibody15 for the antibody-based capture of arginine-GlcNAcylated peptides to perform proteome-wide assessment of Arg-GlcNAcylation mediated by NleB1, as described previously16. We unexpectedly observed glycosylation of E. coli glutathione synthetase (GshB) on arginine residue R256 as the most abundant Arg-GlcNAcylated peptide enriched from samples, followed by the known human NleB1 target FADD16 (Supp. Table 1).
Because C. rodentium encodes only one copy of NleB, while most EHEC strains encode two copies (NleB1 and NleB2), we subsequently attempted to reproduce our initial findings using C. rodentium NleB and GshB using in vivo4 or in vitro17 studies to investigate glycosylation within this protein. Using these assays, we observed that GshB is exclusively modified on R256, with this glycosylation event absent in GshB(256 A) expressing strains [Fig. 1A; -log10(p-value) of 1.77 and 2.78 from in vivo and in vitro assays respectively, Supp. Table 2]. To further test the localization of the glycosylation, in vivo glycosylated GshB was subjected to Electron-Transfer/Higher-Energy Collision Dissociation (EThcD) fragmentation, confirming the attachment of the GlcNAc residue to R256 (Fig. 1B). These data support the notion that the glutathione synthetase GshB from the attaching/effacing pathogens EHEC and C. rodentium is glycosylated at R256 under in vivo conditions.
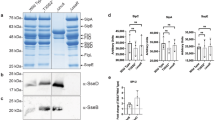
NleB glycosylates GshB R256. (A) Heatmap of Z-scored ion intensities of GshB peptides demonstrates that glycosylated R256 is observed only within WT GshB samples in both in vivo and in vitro glycosylation assays. The abundance of non-glycosylated peptides 109GTLIVNKPQSLRDCNEK125, 88DPPFDTEFIYATYILERAEEK108 and 145AQLKAFWEK153 are unaltered across assays (B) EThcD spectra of the in vivo glycosylated GshB peptide 254IARQIGPTLK263 confirms glycosylation is localized to R256. (C) Western blot analysis of in vitro GshB glycosylation assays. (D) Western blot analysis of in vivo GshB glycosylation assays; Sup, culture supernatant; Pel, bacterial lysate.
To corroborate our mass spectrometry data, we conducted in vitro glycosylation assays17 with the Anti-R-GlcNAc monoclonal antibody15 by expressing recombinant forms of wild-type (WT) GshB or GshB(256 A). NleB glycosylated the WT, but not the R256A GshB mutant (Fig. 1C), consistent with our in vivo and in vitro MS assays. Within these assays, FADD was used as a positive control as a known NleB substrate16. The NleB(AAA) mutant, which lacks glycosylation activity1, was used as a negative control.
We then generated a gshB deletion in C. rodentium and complemented this mutant with FLAG-tagged versions of either WT or GshB(R256A). Both FLAG-tagged forms of GshB were isolated from C. rodentium; only WT GshB was glycosylated by the endogenous levels of NleB (Fig. 1D). Neither secretion of GshB nor its glycosylation in the culture medium by NleB was observed, suggesting that NleB glycosylation of GshB occurred within the bacterium.
NleB promotes oxidative stress resistance
Glutathione (GSH) is generated in Gram-negative bacteria in a two-step process by γ-glutamylcysteine synthetase (gshA) and glutathione synthetase (gshB). GshA uses glutamate and cysteine as substrates to catalyze the formation of γ-L-glutamylcysteine. GshB catalyzes GSH production by ligating glycine and γ-L-glutamylcysteine18. Salmonella lacking either enzyme do not produce GSH and exhibit increased susceptibility to oxidative stress19. A Salmonella gshA deletion is hypersensitive to H2O2, and gshA and gshB deletions are hypersensitive to both nitric oxide (NO) and S-nitrosoglutathione (GSNO)19. Similarly, in Pseudomonas aeruginosa, gshA and/or gshB deletions are more sensitive to environmental stress and attenuated for virulence20. In Streptococcus pneumoniae, mutating gshT, an ABC transporter required for GSH import, increased S. pneumoniae sensitivity to superoxide, and was attenuated in a mouse model of infection21. Deleting gshA and gshB from P. aeruginosa attenuates several virulence-associated phenotypes including motility and biofilm formation. GSH was also shown to activate both the P. aeruginosa T3SS and a subset of T6SS genes22. Glutathione binding to the Listeria monocytogenes master regulator PrfA is also critical to the virulence of this intracellular pathogen23.
To determine whether GshB glycosylation by NleB has functional significance in promoting resistance to oxidative stress, we compared the growth rates of bacterial strains in the presence or absence of H2O2. Both the nleB and gshB mutants had a significant growth defect in the presence of H2O2 (Fig. 2A), despite having similar growth rates in the absence of H2O2 (Fig. 2B). The nleB mutant phenotype was complemented by expressing WT nleB on a plasmid, but not by expressing the inactive nleB(AAA) mutant. Mutating the GshB R256 residue to alanine (R256A) had no impact on bacterial growth rates. These data highlight a new role for NleB, namely its requirement for C. rodentium survival in peroxide stress conditions.
Arg-glycosylation enhances GshB activity
We then tested the hypothesis that GshB glycosylation enhances GshB-mediated production of glutathione (GSH), consistent with the bacterial growth phenotypes in the presence of H2O2. We incubated C. rodentium lysates prepared from cultures grown in the presence or absence of 2.4 mM H2O2 with glutathione S-transferase (GST) and 1-chloro-2,4-dinitrobenzene (CDNB) to quantify GSH concentrations in bacterial lysates24. The WT strain produced significantly more GSH (3.4 + 0.9 pmoles/s) than did the nleB mutant (2.3 + 0.6 pmoles/s), irrespective of the presence of H2O2 (Fig. 3A,B). To verify that the growth defect of the nleB-deleted strain in H2O2 was due to insufficient GSH, we supplemented bacterial cultures with 5.0 mM GSH and observed partial restoration of the growth of the nleB mutant in the presence of H2O2 (Fig. 3C).
GSH production from bacterial lysates. (A) Bacterial lysates derived from C. rodentium strains grown in the absence of H2O2 were incubated with 1 mM CDNB and 1 µM GST. Absorbance was recorded at 340 nm every 20 seconds for 10 minutes and OD340 data were converted into GSH concentrations using a GST standard curve. (B) GSH production from C. rodentium strains grown in the presence of H2O2. (C) Complementation of C. rodentium nleB growth in 2.4 mM H2O2 in the presence or absence of 5.0 mM GSH.
We then reconstituted an in vitro GSH production assay using purified, recombinant GshA, GST, as well as GshB that was purified from E. coli BL21(DE3), following its co-expression with either NleB or NleB(AAA) (Fig. 4A). Thus, we aimed to generate either glycosylated GshB (when co-expressed with WT NleB) or unglycosylated GshB [when mutated to R256A and/or when WT GshB was co-expressed with NleB(AAA)] to directly characterize the impact of GshB glycosylation on GshB activity. Glycosylation of GshB on R256 in this assay was confirmed using western blotting, showing that NleB was active when expressed in E. coli BL21(DE3) (Fig. 4B). The GshB enzyme purified from the WT NleB co-expression strain was significantly more active than the GshB enzyme purified from the NleB(AAA) co-expression strain (1.5 + 0.1 vs. 1.0. + 0.1 pmoles/s) (Fig. 4C). NleB had no impact on GSH production catalyzed by GshB(R256A). We obtained similar data when GshB was first purified from E. coli and then subsequently glycosylated by NleB in vitro (Fig. S1).
In vitro GSH assays. (A) Coomassie blue staining of purified proteins used in GSH assays. (B) Western blot analysis of the GshB glycosylation state from GSH assays. (C) GSH production as a function of time from reactions containing either WT or GshB(256A) purified after their co-expression with either WT or NleB(AAA).
We also considered whether Salmonella might glycosylate GshB in vivo. We did not observe such glycosylation under conditions performed similarly to those using C. rodentium (Fig. S2A). However, Salmonella is significantly more resistant to H2O2 than is C. rodentium, due to the presence of redundant catalases and alkyl hydroperoxide reductases25,26,27. Consistent with this, we failed to observe a growth phenotype when we subjected WT Salmonella and all possible combinations of sseK1/K2/K3 mutants to 12.0 mM H2O2, a concentration 5-times greater than that used for C. rodentium experiments (Fig. S2B). These data suggest that the SseK enzymes are not significantly involved in H2O2 resistance by Salmonella.
However, we further considered the possibility that some degree of GshB glycosylation might be observed in vitro using purified recombinant proteins. We especially considered this because of previous findings demonstrating that NleB/SseK substrate specificity may be more broad in vitro as compared to in vivo assays, particularly when the enzyme is present at supraphysiological concentration16. We incubated GshB with all known NleB/SseK orthologs from EHEC, EPEC, C. rodentium, and Salmonella and observed that EHEC NleB1, EPEC NleB1, and Salmonella SseK1 all glycosylated GshB, whereas NleB2, SseK2, and SseK3 did not (Fig. S2C). Whether GshB glycosylation by EHEC and/or EPEC NleB1 may affect oxidative stress resistance in these attaching/effacing pathogens awaits further experimentation.
Although no chaperone for NleB has been identified, we also considered whether the presence of CesT, a multi-cargo chaperone that interacts with at least 9 other effectors28, might affect NleB activity. We especially considered this possibility because effector-binding and secretion activities are separable for CesT29. However, we did not observe any impact on NleB activity in a C. rodentium cesT mutant (Fig. S2A), nor did we observe an interaction between NleB and CesT, as assessed using affinity chromatography (data not shown). Thus, NleB activity appears to be independent of CesT.
Discussion
NleB is an arginine glycosyltransferase that modifies several host proteins, leading to inactivation of the host pro-inflammatory response mediated by NF-κB4,6. Here we discovered that NleB also plays a significant role in bacterial survival in oxidative stress conditions by glycosylating GshB to enhance GSH production. In this case, rather than inactivating the NleB substrates, as is seen with the mammalian targets, Arg-GlcNAcylation significantly activates the bacterial GshB enzyme. It remains to be determined why GshB glycosylation enhances its enzyme activity. The glycosylated R256 is distant from the active site (Fig. S2B), as inferred from the E. coli GshB structure [PDB 1GSA;30]. Possible mechanisms could include changes in GshB oligomerization state, a conformational change of the GshB active site, or enhanced affinity for/localization with GshA.
GshB is an abundant cytoplasmic protein31. In our assays, we observed a 50–100% increase in GshB activity (Figs. 3–4), despite only a relatively low degree of overall GshB Arg-GlcNAcylation (occupancy of ~5%, Supp. Table 1). It is therefore possible that Arg-GlcNAcylation may increase GshB activity by as much as 10-fold. Formal testing of this hypothesis awaits the complete separation of glycosylated from unglycosylated forms of GshB.
NleB/SseK activities in the host cell have been characterized for their inhibition of proteins involved in the innate immune response1,2,3,4,6. Previous work with Ear-P showed that Arg-rhamnosylation activates EF-P in the bacterial cytosol13, to avoid ribosome stalling during the synthesis of poly-proline regions32. It is now clear from our work that Arg-GlcNAcylation also provides an important activating function, namely the NleB-mediated activation of GshB through R256 glycosylation. The abundance and functional significance of cytoplasmic bacterial glycoproteins may be a fruitful area for future studies.
Regardless of the specific activating mechanism, these data represent, to our knowledge, the first example of a T3SS effector functioning within both the bacterium and within the host cell. It is possible that other C. rodentium proteins are glycosylated by NleB to provide the organism a means by which to integrate T3SS activation and effector synthesis with bacterial physiological processes such as transcriptional regulation, flagellar biosynthesis, and quorum sensing. It is also possible that other T3SS effectors are substrates of NleB, and that other T3SS effectors with enzymatic activities are active within the bacterium, a concept similar to the role of metaeffectors playing important regulatory roles in Legionella pneumophila biology33. The extent to which other effectors with enzymatic activities may be active within the bacterium may emerge as a new area of investigation.
Materials and Methods
Strains and molecular cloning
The plasmids and strains used in this study are listed in Table 1. WT nleB (C. rodentium) and its derivative DAD221–223/AAA were cloned into pET42a. FADD was cloned into pET15a. WT gshB and its derivative gshB R256A, as well as gshA, were cloned in pET28a using ABC cloning34. The C. rodentium gshB deletion was generated using lambda red recombination35.
Protein purification
Proteins were expressed from E. coli BL21 (DE3) and induced with 0.5 mM IPTG when cultures reached an OD600 of 0.4. Cells were grown for an additional 4 h at 37 °C and then harvested using centrifugation. Cell pellets were resuspended in 1/40th culture volume of 50 mM NaH2PO4 pH 8.0 supplemented with 0.5 mg/ml lysozyme and protease inhibitor cocktails (Thermo Scientific). Bacterial suspensions were incubated on ice for 30 min with occasional shaking, at which time an equal volume 50 mM NaH2PO4 pH 8.0, 2 M NaCl, 8 mM imidazole, 20% glycerol, 2% Triton X-100 was added for additional incubation on ice for 30 min. Cell lysates were sonicated, centrifuged, and combined with 2 ml Ni-NTA slurry (Qiagen) for 1 h at 4 °C with gentle rotation. The mixture was loaded on a Poly-Prep Chromatography Column (Bio-Rad) and washed in 50 mM NaH2PO4 pH 8.0, 600 mM NaCl, 60 mM imidazole, 10% glycerol). Proteins were eluted in 50 mM NaH2PO4 pH 8.0, 600 mM NaCl, 250 mM imidazole, 10% glycerol and then dialyzed into the same buffer lacking imidazole17.
Enrichment of arginine-glycosylated peptides from cell lysates
HEK293T cells were grown in DMEM to 80% confluency and then infected with EHEC O111:NM R82F236 at a multiplicity of infection of 1.0 for 16 h. Infected cells were washed three times in ice-cold PBS and lysed by scraping with ice-cold guanidinium chloride lysis buffer (6 M GdmCl, 100 mM Tris pH 8.5, 10 mM TCEP, 40 mM 2-Chloroacetamide) on a bed of ice. Lysates were collected and boiled at 95 °C for 10 minutes with shaking at 2,000 rpm to shear DNA and inactivate protease activity. Lysates were then cooled for 10 minutes on ice and then boiled again at 95 °C for 10 minutes with shaking at 2,000 rpm. Protein samples (2 mg) were acetone precipitated and dried protein pellets were resuspended in 6 M urea, 2 M thiourea, 40 mM NH4HCO3 and reduced/alkylated prior to digestion with Lys-C (1/200 w/w) and then with trypsin (1/50 w/w) overnight as previously described37. Digested samples were acidified to a final concentration of 0.5% formic acid and desalted with 50 mg tC18 SEP-PAK (Waters Corporation). tC18 SEP-PAKs were conditioned with buffer B (80% ACN, 0.1% formic acid), washed with 10 volumes of Buffer A* (0.1% TFA, 2% ACN), sample loaded, column washed with 10 volumes of Buffer A* and bound peptides eluted with buffer B then dried.
Arg-GlcNAc peptide affinity purification was performed as previously described16. Protein A/G plus Agarose beads (Santa Cruz, Santa Cruz CA) were washed with immunoprecipitation buffer (IAP, 10 mM Na2HPO4, 50 mm NaCl, 50 mM MOPS, pH 7.2) and rotated overnight with 10 μg of anti-Arg-GlcNAc antibody (ab195033, Abcam) at 4 °C. Coupled anti-Arg-GlcNAc beads were then washed with 100 mM sodium borate (pH 9) to remove non-bound proteins and cross-linked for 30 minutes of rotation using 20 mM dimethyl pimelimidate in 100 mM HEPES, pH 8.0. Cross-linking was quenched by washing beads three times with 200 mM ethanolamine, pH 8.0 and then rotating the beads in an additional 1 ml of 200 mM ethanolamine, pH 8.0 for 2 hours at 4 °C. Purified peptides were resuspended in 1 ml IAP buffer and peptide lysates were then added to the prepared cross-linked anti-Arg-GlcNAc antibody beads and rotated for 3 hours at 4 °C. Antibody beads were centrifuged at 3,000 g for 2 minutes at 4 °C and the unbound peptide lysates collected. Antibody beads were then washed with ice-cold IAP buffer and Arg-GlcNAc peptides eluted using two rounds of acid elution. For each elution round, 100 μl of 0.2% TFA was added and antibody beads allowed to stand at room temperature with gentle shaking every minute for 10 minutes. Peptide supernatants were collected and desalted using C18 stage tips38 before being dried down and stored until LC-MS analysis.
SP3 on-bead Lys-C digestion of purified GshB
Purified tagged GshB was cleaned up using SP3 based purification according to previous protocols39. Samples were first denatured and reduced using 1% SDS, 10 mM DTT, 100 mM HEPES by boiling at 95 °C, 1,000 rpm for 10 minutes. Samples were then cooled and alkylated with 40 mM 2-chloroacetamide (CAA) for 1 hour at RT in the dark. The alkylation reactions were then quenched with 40 mM DTT for 10 minutes and then samples precipitated on to SeraMag Speed Beads (GE Healthcare, USA) with ethanol (final concentration 50% v/v). Samples were shaken for 10 minutes to allow complete precipitation onto beads and then washed three times with 80% ethanol. The beads were resuspended in 100 mM ammonium bicarbonate containing 1 μg lys-C 1/50 (w/w) and digested overnight at 37 °C. Samples were centrifuged at 14,000 g for 5 minutes to pellet the beads and the supernatant collected and desalted using C18 stage tips before being dried for LC-MS analysis.
Identification of arginine-glycosylated affinity enriched peptides and his-tagged proteins using reversed phase LC-MS
Purified peptides were resuspended in Buffer A* and separated using a two-column chromatography setup composed of a PepMap100 C18 20 mm × 75 μm trap and a PepMap C18 500 mm × 75 μm analytical column (Thermo Fisher Scientific)16. Samples were concentrated onto the trap column at 5 μl/min for 5 minutes and infused into either an Orbitrap Fusion Lumos Tribrid Mass Spectrometer (Thermo Fisher Scientific) for the analysis of enriched Arg-GlcNAc peptides and PRM analysis of Arg-GlcNAcylated GshB or an Orbitrap Elite (Thermo Fisher Scientific) for the comparison of Arg-GlcNAcylation levels within purified GshB. Gradients (120 minutes) were run by altering the buffer composition from 1% buffer B to 28% B over 90 minutes, then from 28% B to 40% B over 10 minutes, then from 40% B to 100% B over 2 minutes, the composition was held at 100% B for 3 minutes, and then dropped to 3% B over 5 minutes and held at 3% B for another 10 minutes for enriched Arg-GlcNAc modified peptides and comparison of Arg-GlcNAcylation levels within purified GshB. For 120-minute gradients, the Lumos and Elite Mass Spectrometers were operated in a data-dependent mode automatically switching between the acquisition of a single Orbitrap MS scan HCD or CID fragmentation. For parallel reaction monitoring (PRM) experiments, the known Lys-C Arg-modified peptide of GshB (IARQIGPTLKEK) was monitored using EThcD fragmentation targeting the predicted m/z for the +2 (m/z 650.2835) and +3 charge (m/z 488.9248) states over a 95 minute gradient, altering the buffer composition from 1% buffer B to 28% B over 60 minutes, then from 28% B to 40% B over 10 minutes, then from 40% B to 100% B over 2 minutes, held at 100% B for 3 minutes, and then dropped to 3% B over 5 minutes and held at 3% B for another 15 minutes.
Mass spectrometry data analysis
Identification of Arg-glycosylated peptides was accomplished using MaxQuant (v1.5.3.30)40. Searches were performed against E. coli O127:H6 strain E2348/69 (Uniprot proteome id UP000008205- E. coli O127:H6 strain E2348/69/EPEC, downloaded 28-07-2014, 4,595 entries) or C. rodentium ICC168 (Uniprot proteome id UP000001889- C. rodentium strain ICC168 downloaded 12/12/2016) proteomes depending on the samples, with carbamidomethylation of cysteine set as a fixed modification. Searches were performed with Trypsin or Lys-C cleavage specificity depending on the experiment. Two miscleavage events were allowed, as well as the variable modifications of oxidation of methionine, N-Acetylhexosamine addition to arginine (Arg-GlcNAc) and acetylation of protein N-termini. Precursor mass tolerance was set to 20 parts-per-million (ppm) for the first search and 10 ppm for the main search, with a maximum false discovery rate (FDR) of 1.0% set for protein and peptide identifications. The Match Between Runs option was enabled with a precursor match window set to 2 minutes and an alignment window of 10 minutes. For label-free quantitation, the MaxLFQ option within Maxquant41 was enabled, in addition to the re-quantification module. Peptide outputs were processed using the Perseus (v1.4.0.6)42 analysis environment to remove reverse matches and common protein contaminants with missing values imputed and the peptide intensities z-scored. MS/MS annotations were undertaken using the Interactive Peptide Spectral Annotator43. Mass spectrometry proteomics data were deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD015752.
Glycosylation assays
In vitro glycosylation assays were performed as described previously17 using 200 nM of NleB1 or its orthologs with 1 μM of either WT GshB or GshB R256A in 50 mM Tris-HCl pH 7.4, 1 mM UDP-GlcNAc, 10 mM MnCl2, and 1 mM DTT. After 2 h incubation at RT, samples were subjected to western blotting using an anti-R-GlcNAc monoclonal antibody (Abcam). In vivo glycosylation assays were performed using C. rodentium and S. enterica strains electroporated with plasmids expressing His-GshB or His-GshB(R256A). Transformed bacteria were grown overnight in the presence of 0.5 mM IPTG, harvested using centrifugation, and then His-tagged proteins were purified as described above, and then subjected to western blotting using an anti-R-GlcNAc monoclonal antibody (Abcam).
Bacterial growth assays
Overnight cultures were used to inoculate 50 ml of LB medium. H2O2 (2.4 mM for C. rodentium or 12.0 mM for S. enterica) was added to cultures when they reached an OD600 of 0.3 and bacterial growth was monitored for 10–16 h at 37 °C.
GSH quantification from bacterial lysates
Bacteria were harvested by centrifugation, lysed in 50 mM Tris-HCl pH 7.4 supplemented with 0.5 ug/ml lysozyme, and recentrifuged. The supernatant was incubated with 1 mM CDNB and 1 µM GST. Absorbance was recorded at 340 nm every 20 seconds for 10 minutes and OD340 data were converted into molar concentrations of GSH using a GST standard curve24.
In-vitro GSH assays
His-GshB or His-GshB(R256A) were purified over Ni-NTA slurries after they were respectively co-expressed with either NleB-FLAG or NleB(AAA)-FLAG in E. coli BL21(DE3) cells. An in vitro GSH assay was then prepared by first incubating GshA (1 µM) with 5 mM glycine, 5 mM cysteine, and 5 mM glutamate in a reaction buffer containing 50 mM Tris HCl pH 7.4, 1 mM DTT, 1 mM MgCl2, 1 mM ATP, and 3% DMSO. After 1 h incubation at 37 °C, the purified GshB proteins (50 nM), along with 100 nM GST and 1 mM CDNB were added, and glutathione formation was monitored by reading the absorbance at 340 nm as a function of time.
Statistical analyses
Bacterial growth assays were analyzed using non-linear regression followed by Dunn’s multiple comparison testing. GSH assays were analyzed using linear regression. p-values <0.05 were considered significant.
References
Gao, X. et al. NleB, a bacterial effector with glycosyltransferase activity, targets GAPDH function to inhibit NF-kappaB activation. Cell host microbe 13, 87–99, https://doi.org/10.1016/j.chom.2012.11.010 (2013).
Li, S. et al. Pathogen blocks host death receptor signalling by arginine GlcNAcylation of death domains. Nat. 501, 242–246, https://doi.org/10.1038/nature12436 (2013).
Pearson, J. S. et al. A type III effector antagonizes death receptor signalling during bacterial gut infection. Nat. 501, 247–251, https://doi.org/10.1038/nature12524 (2013).
Ding, J. et al. Structural and Functional Insights into Host Death Domains Inactivation by the Bacterial Arginine GlcNAcyltransferase Effector. Molecular cell 74, 922-935.e926, https://doi.org/10.1016/j.molcel.2019.03.028 (2019).
Esposito, D. et al. Structural basis for the glycosyltransferase activity of the Salmonella effector SseK3. J. Biol. Chem. 293, 5064–5078, https://doi.org/10.1074/jbc.RA118.001796 (2018).
Park, J. B. et al. Structural basis for arginine glycosylation of host substrates by bacterial effector proteins. Nat. Commun. 9, 4283, https://doi.org/10.1038/s41467-018-06680-6 (2018).
Deng, W. et al. Assembly, structure, function and regulation of type III secretion systems. Nat. Rev. Microbiol. 15, 323–337, https://doi.org/10.1038/nrmicro.2017.20 (2017).
Wagner, S. et al. Bacterial type III secretion systems: a complex device for the delivery of bacterial effector proteins into eukaryotic host cells. FEMS microbiology letters 365, https://doi.org/10.1093/femsle/fny201 (2018).
Feldman, M. F. & Cornelis, G. R. The multitalented type III chaperones: all you can do with 15 kDa. FEMS microbiology Lett. 219, 151–158, https://doi.org/10.1016/s0378-1097(03)00042-9 (2003).
Young, C. & Nester, E. W. Association of the virD2 protein with the 5’ end of T strands in Agrobacterium tumefaciens. J. Bacteriol. 170, 3367–3374, https://doi.org/10.1128/jb.170.8.3367-3374.1988 (1988).
Pelczar, P., Kalck, V., Gomez, D. & Hohn, B. Agrobacterium proteins VirD2 and VirE2 mediate precise integration of synthetic T-DNA complexes in mammalian cells. EMBO Rep. 5, 632–637, https://doi.org/10.1038/sj.embor.7400165 (2004).
Kusmierek, M. et al. A bacterial secreted translocator hijacks riboregulators to control type III secretion in response to host cell contact. PLoS Pathog. 15, e1007813, https://doi.org/10.1371/journal.ppat.1007813 (2019).
Lassak, J. et al. Arginine-rhamnosylation as new strategy to activate translation elongation factor P. Nat. Chem. Biol. 11, 266–270, https://doi.org/10.1038/nchembio.1751 (2015).
Lassak, J., Koller, F., Krafczyk, R. & Volkwein, W. Exceptionally versatile - arginine in bacterial post-translational protein modifications. Biol. Chem. 400, 1397–1427, https://doi.org/10.1515/hsz-2019-0182 (2019).
Pan, M. et al. Synthesis of and specific antibody generation for glycopeptides with arginine N-GlcNAcylation. Angew. Chem. Int. Ed. Engl. 53, 14517–14521, https://doi.org/10.1002/anie.201407824 (2014).
Scott, N. E. et al. The bacterial arginine glycosyltransferase effector NleB preferentially modifies Fas-associated death domain protein (FADD). J. Biol. Chem. 292, 17337–17350, https://doi.org/10.1074/jbc.M117.805036 (2017).
El Qaidi, S. et al. NleB/SseK effectors from Citrobacter rodentium, Escherichia coli, and Salmonella enterica display distinct differences in host substrate specificity. J. Biol. Chem. 292, 11423–11430, https://doi.org/10.1074/jbc.M117.790675 (2017).
Lushchak, V. I. Glutathione homeostasis and functions: potential targets for medical interventions. J. amino acids 2012, 736837, https://doi.org/10.1155/2012/736837 (2012).
Song, M. et al. Low-molecular-weight thiol-dependent antioxidant and antinitrosative defences in Salmonella pathogenesis. Mol. microbiology 87, 609–622, https://doi.org/10.1111/mmi.12119 (2013).
Wongsaroj, L. et al. Pseudomonas aeruginosa glutathione biosynthesis genes play multiple roles in stress protection, bacterial virulence and biofilm formation. PLoS one 13, e0205815, https://doi.org/10.1371/journal.pone.0205815 (2018).
Potter, A. J., Trappetti, C. & Paton, J. C. Streptococcus pneumoniae uses glutathione to defend against oxidative stress and metal ion toxicity. J. Bacteriol. 194, 6248–6254, https://doi.org/10.1128/jb.01393-12 (2012).
Zhang, Y. et al. Glutathione Activates Type III Secretion System Through Vfr in Pseudomonas aeruginosa. Frontiers in Cellular and Infection Microbiology 9, https://doi.org/10.3389/fcimb.2019.00164 (2019).
Reniere, M. L. et al. Glutathione activates virulence gene expression of an intracellular pathogen. Nat. 517, 170–173, https://doi.org/10.1038/nature14029 (2015).
Habig, W. H., Pabst, M. J. & Jakoby, W. B. Glutathione S-transferases. The first enzymatic step in mercapturic acid formation. J. Biol. Chem. 249, 7130–7139 (1974).
Hebrard, M., Viala, J. P., Meresse, S., Barras, F. & Aussel, L. Redundant hydrogen peroxide scavengers contribute to Salmonella virulence and oxidative stress resistance. J. Bacteriol. 191, 4605–4614, https://doi.org/10.1128/JB.00144-09 (2009).
Horst, S. A. et al. Thiol peroxidase protects Salmonella enterica from hydrogen peroxide stress in vitro and facilitates intracellular growth. J. Bacteriol. 192, 2929–2932, https://doi.org/10.1128/JB.01652-09 (2010).
Karash, S., Liyanage, R., Qassab, A., Lay, J. O. Jr. & Kwon, Y. M. A Comprehensive Assessment of the Genetic Determinants in Salmonella Typhimurium for Resistance to Hydrogen Peroxide Using Proteogenomics. Sci. Rep. 7, 17073, https://doi.org/10.1038/s41598-017-17149-9 (2017).
Thomas, N. A., Deng, W., Baker, N., Puente, J. & Finlay, B. B. Hierarchical delivery of an essential host colonization factor in enteropathogenic Escherichia coli. J. Biol. Chem. 282, 29634–29645, https://doi.org/10.1074/jbc.M706019200 (2007).
Ramu, T. et al. A novel C-terminal region within the multicargo type III secretion chaperone CesT contributes to effector secretion. J. Bacteriol. 195, 740–756, https://doi.org/10.1128/JB.01967-12 (2013).
Hara, T., Kato, H., Katsube, Y. & Oda, J. A pseudo-michaelis quaternary complex in the reverse reaction of a ligase: structure of Escherichia coli B glutathione synthetase complexed with ADP, glutathione, and sulfate at 2.0A resolution. Biochem. 35, 11967–11974, https://doi.org/10.1021/bi9605245 (1996).
Lopez-Campistrous, A. et al. Localization, annotation, and comparison of the Escherichia coli K-12 proteome under two states of growth. Mol. Cell Proteom. 4, 1205–1209, https://doi.org/10.1074/mcp.D500006-MCP200 (2005).
Ude, S. et al. Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Sci. 339, 82–85, https://doi.org/10.1126/science.1228985 (2013).
Urbanus, M. L. et al. Diverse mechanisms of metaeffector activity in an intracellular bacterial pathogen, Legionella pneumophila. Mol. Syst. Biol. 12, 893, https://doi.org/10.15252/msb.20167381 (2016).
Qaidi, S. E. & Hardwidge, P. R. ABC cloning: An efficient, simple, and rapid restriction/ligase-free method. MethodsX 6, 316–321, https://doi.org/10.1016/j.mex.2019.02.007 (2019).
Datsenko, K. A. & Wanner, B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. U S Am. 97, 6640–6645, https://doi.org/10.1073/pnas.120163297 (2000).
Karmali, M. A. et al. Association of genomic O island 122 of Escherichia coli EDL 933 with verocytotoxin-producing Escherichia coli seropathotypes that are linked to epidemic and/or serious disease. J. Clin. Microbiol. 41, 4930–4940, https://doi.org/10.1128/jcm.41.11.4930-4940.2003 (2003).
Scott, N. E. et al. Simultaneous glycan-peptide characterization using hydrophilic interaction chromatography and parallel fragmentation by CID, higher energy collisional dissociation, and electron transfer dissociation MS applied to the N-linked glycoproteome of Campylobacter jejuni. Mol. Cell Proteom. 10, M000031–MCP000201, https://doi.org/10.1074/mcp.M000031-MCP201 (2011).
Rappsilber, J., Mann, M. & Ishihama, Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906, https://doi.org/10.1038/nprot.2007.261 (2007).
Hughes, C. S. et al. Single-pot, solid-phase-enhanced sample preparation for proteomics experiments. Nat. Protoc. 14, 68–85, https://doi.org/10.1038/s41596-018-0082-x (2019).
Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372, https://doi.org/10.1038/nbt.1511 (2008).
Cox, J. et al. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell Proteom. 13, 2513–2526, https://doi.org/10.1074/mcp.M113.031591 (2014).
Tyanova, S. et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740, https://doi.org/10.1038/nmeth.3901 (2016).
Brademan, D. R., Riley, N. M., Kwiecien, N. W. & Coon, J. J. Interactive Peptide Spectral Annotator: A Versatile Web-based Tool for Proteomic Applications. Mol. Cell Proteom. 18, S193–S201, https://doi.org/10.1074/mcp.TIR118.001209 (2019).
Luperchio, S. A. et al. Citrobacter rodentium, the causative agent of transmissible murine colonic hyperplasia, exhibits clonality: synonymy of C. rodentium and mouse-pathogenic Escherichia coli. J. Clin. Microbiol. 38, 4343–4350 (2000).
Deng, W. et al. Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc. Natl Acad. Sci. U S Am. 101, 3597–3602, https://doi.org/10.1073/pnas.0400326101 (2004).
El Qaidi, S. et al. High-Throughput Screening for Bacterial Glycosyltransferase Inhibitors. Front. Cell Infect. Microbiol. 8, 435, https://doi.org/10.3389/fcimb.2018.00435 (2018).
Acknowledgements
The project described was supported by grant number AI127973 (PRH) from the National Institute of Allergy and Infectious Diseases (NIAID). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIAID. This work was also supported by National Health and Medical Research Council of Australia (NHMRC) project grants awarded to NES (APP1100164). We thank the Melbourne Mass Spectrometry and Proteomics Facility of The Bio21 Molecular Science and Biotechnology Institute at The University of Melbourne for the support of mass spectrometry analysis.
Author information
Authors and Affiliations
Contributions
S.E., N.S., M.P.H. and S.W. performed the experiments N.S., B.V.G. and P.R.H. analyzed the data, wrote the main manuscript text and prepared figures. All authors reviewed the data.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
El Qaidi, S., Scott, N.E., Hays, M.P. et al. An intra-bacterial activity for a T3SS effector. Sci Rep 10, 1073 (2020). https://doi.org/10.1038/s41598-020-58062-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-58062-y
This article is cited by
-
Salmonella T3SS effector SseK1 arginine-glycosylates the two-component response regulator OmpR to alter bile salt resistance
Scientific Reports (2023)
-
Arginine glycosylation regulates UDP-GlcNAc biosynthesis in Salmonella enterica
Scientific Reports (2022)
-
Catalytic DxD motif caged in Asx-turn and Met–aromatic interaction attenuates the pathogenic glycosylation of SseK2/NleB2 effectors
Scientific Reports (2022)
-
Burkholderia PglL enzymes are Serine preferring oligosaccharyltransferases which target conserved proteins across the Burkholderia genus
Communications Biology (2021)
-
Arginine glycosylation enhances methylglyoxal detoxification
Scientific Reports (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.