Abstract
The gene GIGANTEA (GI) appeared early in land plants. It is a single copy gene in most plants and is found in two to three copies in Solanaceae. We analyzed the silencing of one GI copy, Petunia hybrida GI1 (PhGI1), by hairpin RNAs in Petunia in order to gain knowledge about its range of functions. Decreased transcript levels of PhGI1 were accompanied also by a reduction of PhGI2. They were further associated with increased time period between two consecutive peaks for PhGI1 and CHANEL (PhCHL), the orthologue of the blue light receptor gene ZEITLUPE (ZTL), confirming its role in maintaining circadian rhythmicity. Silenced plants were bigger with modified internode length and increased leaf size while flowering time was not altered. We uncovered a new function for PhGI1 as silenced plants showed reduction of flower bud number and the appearance of two flower buds in the bifurcation point, were normally one flower bud and the inflorescence meristem separate. Furthermore, one of the flower buds consistently showed premature flower abortion. Flowers that developed fully were significantly smaller as a result of decreased cell size. Even so the circadian pattern of volatile emission was unchanged in the silenced lines, flowers emitted 20% less volatiles on fresh weight basis over 24 hours and showed changes in the scent profile. Our results indicate a novel role of PhGI1 in the development of reproductive organs in Petunia. PhGI1 therefore represses growth in vegetative plant parts, maintains the typical cymose inflorescence structure, and inhibits premature flower abortion.
Similar content being viewed by others
Introduction
The evolution of land plants has included amongst other adaptations the increase in complexity of the circadian clock. Predictable changes in the environment, as light and temperature, are anticipated by the plant circadian clock, which allows them to adjust their developmental and physiological traits. Most detailed studies on plant circadian clock have been performed in Arabidopsis thaliana1. The plant circadian clock is based on a set of genes forming several overlapping loops interacting with each other via transcriptional and post-translational activation and repression2. Based on the time of the day when the mRNA of the gene shows its expression maximum, the genes included in this oscillator have been classified as the morning loop, midday or core loop and evening loop3. In Arabidopsis, LATE ELONGATED HYPOCOTYL (LHY) and CIRCADIAN CLOCK ASSOCIATED 1 (CCA1), two MYB transcription factors, form the central circadian oscillator complex, together with PSEUDO RESPONSE REGULATOR 1 (PRR1), better known as TIMING OF CAB1 (TOC1). PSEUDO-RESPONSE REGULATOR 9 (PRR9) and 7 (PRR7) form the morning loop genes and the evening complex is formed by the three proteins EARLY FLOWERING 3 and 4 (ELF3 and ELF4) and LUX ARHYTHMO (LUX). These clock genes are interconnected via negative autoregulatory feedback loops, meaning that they reciprocally regulate each other1,4,5,6. Light input is received by ZEITLUPE (ZTL), a gene containing an F-box domain and a blue-light sensing domain, which sustains a normal circadian period through proteasome-dependent degradation of the central clock protein TOC17. The stabilization of the ZTL protein in turn is obtained through GIGANTEA (GI), a protein with chaperone activity that facilitates ZTL maturation into an active form8,9.
Studies in Arabidopsis thaliana and other species revealed that the complex wiring of the oscillator network includes an interplay with hormone signaling10,11,12, cell division and expansion13,14, primary metabolism15, abiotic stress response16, the expression of seed storage proteins17, biomass production18,19,20, flower orientation19 and flower scent emission21,22,23,24.
The genetic structure of the circadian clock in the picoeukaryote Ostreococcus includes two genes, a TOC1 and a LHY ortholog25,26. A protein comprising a LOV domain and a histidine kinase appears to function as an entry for light cues27. Relative to these two clock genes, other clock genes such as GI appear later in evolution and are present in Marchantia but not in Physcomitrella patens28. The gene GI encodes a protein that is not fully characterized. It has important functions in plant development including a conserved role in floral transition in Marchantia and Arabidopsis29,30. It plays a role in control of circadian rhythm in Arabidopsis31. Furthermore it coordinates both photoperiod-mediated and independent flowering32,33, growth cessation34, carbohydrate metabolism35, salt tolerance36 and cold stress response37. GI also affects hypocotyl growth in Arabidopsis38,39, and this function is related to gibberellin signaling, as SPINDLY (SPY) protein, a negative regulator of gibberellin signaling in Arabidopsis and an inhibitor of hypocotyl elongation, interacts with GI protein39. Loss of function of GI results in long petioles, tall plant height and many rosette leaves, together with delayed flowering time.
Flower formation in Petunia involves the activity of the flower-meristem-identity genes PETUNIA FLOWERING GENE (PFG) and ALF (ABERRANT LEAF AND FLOWER), the Petunia orthologue of LEAFY of Arabidopsis, which induce the floral fate in the lateral shoot meristem40,41. The typical determinate inflorescence architecture in Petunia is characterized by a bifurcation of the inflorescence meristem, one terminating into a floral meristem, the other maintaining inflorescence identity and repeating the cymose floral pattern. A few mutants show altered architectures, including extra petals (exp), which forms a single terminal flower40 and the mutants alf and double top (dot), a homolog of UNUSUAL FLORAL ORGANS (UFO) from Arabidopsis, where the apical floral meristems convert into inflorescence meristems that do not produce flowers42. Overexpression of DOT leads to the production of a solitary flower43. Another gene that determines Petunia inflorescence architecture is EVERGREEN (EVG) involved in the activation of DOT, the initiation of the floral identity in the apical meristem as well as lateral inflorescence shoot development44. Once the floral program is activated, angiosperm flowers form concentric whorls of organs that include sepals, petals, stamens and carpel and this organ specification relies on the combinatorial genetic function of the organ identity genes according to the Petunia ABCD model45.
GI generally has remained as a single copy gene in most species. Based on the comparison of the Petunia genomes with other Solanaceae, it has been shown that the circadian clock comprises a different set of genes, including GI, which in some cases is duplicated or triplicated. The GI gene is present in two copies in P. axillaris and three in P. inflata. It was hypothesized that some duplicated clock genes may have undergone a subfunctionalization or redeployment46.
In this work, we have characterized one of the GI orthologues from Petunia hybrida, GI1, by creating loss of function plants, using hairpin RNA constructs of PhGI1. Our results demonstrate novel roles of GI1 during flowering, consisting in the promotion of flower initiation and flower maturation, the maintenance of cymose inflorescence structure as well as a control over the species-specific VOC profile.
Results
Silencing of PhGI1 has minor effects on clock gene expression and rhythmicity
We have previously shown that PhGI1 and PhGI2 have similar expression pattern under a 12:12 Light:Dark photoperiod (12:12 LD)47. But we also found that under free running conditions of 12:12 DD, it has a significant change in expression. Thus, we analyzed the expression of PhGI1 as well as PhGI2 under long photoperiods of 16:8 LD and continuous darkness.
The expression pattern of PhGI1 and PhGI2 in wildtype leaves is shown in Fig. 1a. Both PhGI1 and PhGI2 showed a similar pattern of oscillation with peak expression at 9 hours of light (ZT9). We compared the expression levels of PhGI1 during 24 hours, measured in 3-hour intervals, and they were inferior to those of PhGI2 at most time points. Indeed, at ZT9, PhGI2 expression was double than PhGI1. The expression of PhGI had been determined previously using an EST from Petunia24. A DNA alignment of PhGI1, PhGI2 and the previously reported PhGI showed that this EST (FN03636) corresponds to PhGI2 (Fig. S1).
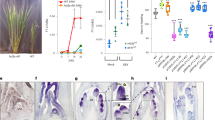
Expression profile during 24 hours of (a) PhGI1 and PhGI2 in wild type plants and (b) PhGI1, (c) PhGI2, (d) PhCHL, (e) PhLHY, (f) PhTOC and (g) PhELF4 in iRNA::PhGI1 T1 lines 3.7, 4.7, 8.1 compared to expression in the wild type (from ZT 0 to ZT 15 of light and from ZT 15 to ZT 24 of dark). Expression represents the normalized expression NE according to the formula (NE) = 2^-(Ct experimental – Ct normalization). Three samples were analyzed for each time point and error bars indicate the standard deviation. Asterisks indicate statistical significance between wild type and iRNA lines with *P < 0.05; **P < 0.01; ***P < 0.001 according to Student’s T-test.
We silenced PhGI1 by hairpin RNAi and obtained several independent transgenic lines. We selected three for further work, iRNA::PhGI1 3.7, 4.7 and 8.1. As a result of silencing, all iRNA::PhGI1 lines lacked the strong increase in PhGI1 expression at ZT9 shown by wild type plants (Fig. 1b) and expression was significantly downregulated at most time points. On average, the peak levels between wild type and iRNA::PhGI lines differed by a factor of 6.
In order to specifically silence only the PhGI1 without cross-silencing PhGI2, we selected a sequence specific for PhGI, as indicated by the level of similarity according to the sequence alignment (Fig. S2). However, as shown in Fig. 1c, PhGI2 was downregulated to 50% in iRNA::PhGI1 lines compared to the wildtype at peak expression, indicating a certain level of cross-silencing.
Figure 1d–g shows the expression of several circadian genes in the wild type compared to iRNA::PhGI1 lines. The genes include CIRCADIAN CLOCK ASSOCIATED1 (LHY) and TIMING OF CAB EXPRESSION1 (TOC1), belonging to the core midday loop, and EARLY FLOWERING 4 (ELF4) and CHANEL, the ortholog of ZTL in wild type Petunia, belonging to the evening loop.
In Arabidopsis, ZTL messenger RNA is uniformly expressed, but ZTL protein levels oscillate with a threefold change in amplitude. Even so no rhythmic expression of ZTL exists in Arabidopsis and Nicotiana attenuata or PhCHL, respectively, in Petunia hybrida21,22,48,49 a significant rise at ZT9 was observed in Petunia leaves at a 12:12 LD light regime47. Similar to this observation we observed peak expression at midday (ZT6), both in wild type and the silenced lines (Fig. 1d).
The expression profile of PhLHY (Fig. 1e) over a 24 hour period (16:8 LD) followed the typical peak at the end of the dark period reported for a wide range of tissues in Arabidopsis and other plants24,50. This pattern was not altered in the silenced lines of PhGI1.
The expression profile of TOC1 over a 24 hours period (16:8 LD) was characterized by lowest levels during dark period and until midday, followed by an increase to peak expression at 12 hours of light, corresponding to the late afternoon, and a sharp decline towards dark period (Fig. 1f). Peak expression towards the end of the day was also reported for Arabidopsis51, soybean16 or cowpea17. In our case, the expression pattern in iRNA::PhGI1 lines was similar to wildtype plants however, peak expression was advanced by three hours in all three transgenic silenced lines (Fig. 1f).
The gene PhELF4, belonging to the evening loop, showed an identical expression, both concerning pattern and expression level, in wildtype and silenced lines, characterized by a peak expression towards the evening (ZT12), followed by a steady decline towards the end of dark period (Fig. 1g).
The mathematical analysis for circadian oscillation using the JTK_CYCLE algorithm (Table 1) showed that all analyzed clock genes had a rhythmic gene expression pattern, both in wildtype and iRNA::PhGI1 lines. In case of GI2, the ZTL-orthologue CHL, and LHY, a significant shift in phase was observed for one or two of the silenced lines, but lacked consistency over all silenced lines. Concerning changes in the time period between two consecutive peaks, a consistent change in all iRNA::PhGI1 lines was observed for GI1 and ZTL-orthologue CHL, which prolonged from 21 to 24 hours.
We analyzed the effect of continuous dark conditions on the expression of PhGI1 in RNA lines. As shown in Fig. 2, wild type and silenced lines showed the typical high and reduced peak under light towards the afternoon, respectively, but revealed very low basic expression levels during the subsequent continuous darkness. Wild type and silenced lines lost rhythmicity during continuous darkness.
Expression profile in leaves under light and subsequent continuous darkness of (a) PhGI1 and PhGI2 in wild type plants and iRNA::PhGI1 T1 lines 3.7 (b), 4.7 (c) and 8.1 (d). Expression represents the normalized expression NE according to the formula (NE) = 2^-(Ct experimental – Ct normalization). Three samples were analyzed for each time point and error bars indicate the standard deviation. Asterisks indicate statistical significance between wild type and iRNA lines with *P < 0.05; **P < 0.01; ***P < 0.001 according to Student’s T-test.
Altogether we can conclude that a strong silencing of PhGI1 does not have a major effect on the expression pattern or rhythmicity of other clock genes. As GI function is via protein-protein interactions but is not known to be part of a transcriptional complex, it could still have major effects on the protein quantities of PhCHL. Furthermore, the rhythmic expression of PhGI1 appears to depend on photoperiod.
PhGI1 is a negative regulator of vegetative growth
We analyzed the effect of downregulating PhGI1 on vegetative growth. The mean value of the vegetative parameters of transgenic plants from T1 and T2 generation, belonging to 6 independent lines (2, 3, 4, 5, 7 and 8), are given in Table 2. Additionally, the results of three different T1 transgenic plants belonging to the three iRNA::PhGI1 independent lines (3.7, 4.7 and 8.1), are given in Table S1. For the T2 generation we analyzed at least three plants per line. Both generations, grown under long-day conditions, the T1 lines in the growth chamber, and T2 lines in the greenhouse, had significantly longer and broader basal and apical leaves compared to the wild type. Overall, GI1 silenced plants of the three independent lines showed a remarkable modification in leaf area and growth habit (Fig. 3a,b).
Vegetative growth characteristics in iRNA::PhGI1 T1 lines compared to wild type plants under growth chamber conditions of 16 hours light/ 8 hours darkness. (a) From the bottom to the top, basal, medium and apical leaves of three wild type leaves (left) compared to three leaves of PhGI1 3.7, 4.7 and 8.1 lines (right) with the strongest silencing. (b) Growth habit of the transgenic lines compared to the wild type. Wild type plant (left) and iRNA::PhGI1 line 4.7 (right).
In case of T1 generation, all three lines had a denser apical foliar apparatus, characterized by an apparent increase in the foliar volume. The bushier phenotype may result from an average reduction of the internode length of 37.6% in the median plant region and 33.7% in the apical plant region, while basal internode distance was increased by 34.4%, resulting in overall only slightly taller plants (Table 2). A similar, although minor, significant effect on internode length was observed in the T2 generation, resulting in a plant height identical to wildtype. A marked increase in the number of axillary meristems in both generations may also contribute to a bushier phenotype. However, no significant increases in the number of lateral branches or total leaf number was recorded compared to the wildtype (Table 2).
Transgenic lines had a greener appearance in the denser apical regions, while basal leaves were more yellowish (Fig. 3a). This phenotype coincided with a significant decrease in the relative chlorophyll content in basal leaves and a progressive increase in the median and apical ones compared to the wildtype (Table 2).
We can conclude that PhGI1 plays a role in vegetative development with a clear acropetal gradient, as it has opposite effects during early stages of development and middle to late stages.
Flowering time, expressed as the percentage of plants which fully bloomed after rooted shoots were transferred from in vitro jars to pots (in weeks) is given in Fig. 4. iRNA::PhGI1 lines of T1 and T2 generation and wildtype plants flowered contemporaneously, indicating that in contrast to Arabidopsis, PhGI1 does not play a role in floral transition. In general, plants kept in the greenhouse flowered five to eight weeks earlier than those kept at 16:8 LD in the growth chamber corroborating that fluence accelerates floral transition in Petunia.
Percentage of fully open flowers in weeks after transplanting from in vitro culture to substrate of T1 and T2 generation of iRNA::PhGI1 lines compared to wild type plant. T1 lines were grown under growth chamber conditions of 16 hours light/ 8 hours darkness. T2 lines were grown in a greenhouse under natural long-day conditions.
PhGI1 inhibits ectopic flower formation and premature flower senescence
We analyzed floral development in iRNA::PHGI1 plants compared to the wildtype and observed that even though wildtype plants and silenced plants started to flower concurrently (Fig. 4), the silenced plants developed new inflorescences at a slower pace so that the total number of flower buds at the end of flowering period was reduced by 58 and 59% in the T1 and T2 generation, respectively, compared to the wildtype (Tables 3 and S2). Additionally, we found a striking phenotype that we had not seen previously in wildtype plants. We found that at many bifurcation points were the terminal flower and the inflorescence shoot divide, an additional ectopic flower bud appeared (Fig. 5). While the normally positioned flower bud tended to develop to full maturity, the ectopic flower bud appeared to undergo early senescence and aborted. Ectopic flower buds accounted for 40% in T1 and 21% in T2 lines. As a consequence of ectopic flower bud abortion (Figs. 6 and 7) and slower inflorescence development in the transgenic lines, the final percentage of fully developed flowers diminished to 76% and 67% in the T1 and T2 generation, respectively, compared to wild type plants (Table 3).
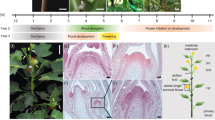
Schematic representation of the petunia inflorescence. Wild type (a) and silenced iRNA::PhGI1 plants of T1 line (c). Side view of a wild type petunia inflorescence (b) and iRNA::PhGI1 plants of T1 line (d–f). Arrows indicate the position and direction of the main and aborted (*) floral meristems at each bifurcation.
Flower size and flower appearance in T1 lines of iRNA::PhGI1 compared to the wild type. Tube length (a) of the flowers of the wild type (left) and transgenic lines (right). (b) Corolla diameter and the abortive flower appearance (extreme right) in T1 lines of iRNA::PhGI1 (right) compared to the wild type (left).
Stages of Petunia flower bud development in wild type and iRNA::PhGI1 T1 lines. Stages (S) 1–7 represent flower development between 27 to 1 days before anthesis and stages (S) 8–11 represent flowers from 0 to 3 days after anthesis. (a) Stages S1 and S2 are given as scanning electron micrographs of the inflorescence apex of Petunia during early stages of development from a wildtype plant (S1) and a plant of iRNA::PhGI1 T1 line (S2). Stigma (St), style (st), ovary (ov), sepal (se), petal (pe), anthers (an). (b) Stages S8–11 of flower buds taken from a position within the plant that develops into a normal flower. (c) Stage S3 of flower buds taken from a position within the plant that develops into an aborted flower.
The differentiation of stamen and carpel tissue could be clearly observed in the aborting flower buds, even so the pale and brownish coloration in iRNA lines indicated an abortion in development, chlorophyll loss and necrosis (Figs. 7 and S3). This early onset of flower senescence occurred well before flowers achieved the normal size of flower opening in Petunia.
The flowers of the silenced lines that fully developed appeared to be smaller with a significant reduction both in the corolla diameter as well as the floral tube length (Table 3, Fig. 6). Cell size in the floral tube and two regions of the corolla, the distal outer zone and a proximal zone near the tube, were significantly reduced (Table 4; Fig. 8), indicating an effect of GI1 silencing over petal cell expansion.
PhGI1 regulates the quantity of volatile emission and fine-tuning of volatile profile
The main VOCs were analyzed in the three different plants with the strongest silencing, belonging to three independent iRNA::PhGI1 lines (3.7, 4.7 and 8.1) and in the wildtype. Data are listed in the Table S3. iRNA::PhGI1 lines showed an average reduction of 20.6% in total VOC emission on the basis of flower fresh weight in grams (Fig. 9a). Figure 9b shows the rhythm of VOC emission during 24 hours in 3 hour intervals, which was similar in case of wildtype flowers and iRNA::PhGI1 lines, with lowest emission towards midday at 6 hours of light and increases towards the end of light period with highest emission during the dark phase. We also observed a change in the relative composition (Figs. 10a–c and Table S4). In both, wildtype plants and iRNA::PhGI1 lines, methyl benzoate was the major volatile with exception of the beginning of light period in iRNA::PhGI1 lines, when this compound contributed with only 30% to the VOC profile. Concerning the relative contribution of other compounds, we found remarkable changes among wildtype plants and iRNA::PhGI1 lines, especially with a high contribution of isoeugenol and ethylbenzoate at certain timepoints (Fig. 10b,c). Results indicate that even so the pattern of total emission during 24 hours is quite conserved, individual VOC compounds may change their emission pattern.
Volatile emission by flowers from wild type and iRNA::PhGI1 T1 lines. Flowers were excised at ZT0. (a) Total VOC emission in wild type flowers compared to iRNA::PhGI1 lines in 24 hours and (b) VOCs emission in three hour intervals during 24 hours. Absolute total emission of VOCs per grams of fresh weight is given as sum of integrated peak area. Asterisks indicate statistical significance between wild type and iRNA lines with *P < 0.05; **P < 0.01; ***P < 0.001 according to Student’s T-test.
Percent emission of volatile organic compounds (VOCs) from wild type flowers and iRNA::PhGI1 T1 lines 3.7, 4.7 and 8.1 Flowers were excised at ZT0. Methyl benzoate (a) and other main VOCs in wildtype flowers (b) and iRNA::PhGI1 lines in three hour intervals during 24 hours. Percentages were calculated based on the integrated peak area divided by flower fresh weight.
Discussion
In this work we have performed a functional analysis of GI1 in Petunia hybrida by means of loss of function using RNAi lines. There are two paralogs in Petunia x hybrida and both PhGI1 and PhGI2 showed a pattern of expression in wildtype plants under a 16:8 LD cycle characterized by an increase towards the afternoon at ZT9 followed by a decrease to very low levels during the entire dark period. This pattern is similar to that observed in Arabidopsis, were under long day conditions (16:8 LD), GI mRNA peaks at ZT 10 and lowest expression levels occur at ZT 030. Other examples of an evening phased expression pattern for GI include cowpea17,52 and soybean16. In Arabidopsis, CCA1 binds to the G1 promoter and reduces its expression, which only rises towards midday, when CCA1 expression is repressed by TOC153. We did not observe a robust circadian rhythmicity under continuous darkness for PhGI1 in the wildtype or the transgenic silenced lines. PhGI1 expression in Petunia therefore diverges from Arabidopsis, were a strong oscillation of GI under conditions of continuous darkness can be observed54. Observations on Petunia hybrida leaves confirm that PhGI2 does not maintain rhythmicity during continuous darkness24, indicating the necessity of light for the correct expression signals for oscillation of the GI paralogs in Petunia.
The specific silencing of a gene for which several paralogs exist within a plant species is very challenging, as it might be difficult to find regions with a sufficient degree of sequence variation. We selected the 3′ untranslated region of PhGI1 and PhGI2, that showed the maximum sequence differences. Even so gene specific sequences were selected for the silencing of PhGI1 in order to avoid cross-silencing with PhGI2, the selected sequence still contained stretches of identical sequences. As a result, we observed a certain level of silencing of PhGI2. While PhGI1 expression levels were down regulated on average 5.6 fold in iRNA::PhGI1 lines, the reduction of PhGI2 in these lines was 2.3 fold compared to wild type. Silencing of non-targeted genes was reported to occur if these targets contain as few as eleven contiguous nucleotides of identity with the siRNA sequence55, which might explain the observed off-target effects in case of these duplicated GI genes. Currently, we cannot determine if the downregulation of PhGI2 is the result of the iRNA::PhGI1 construct or if PhGI1 activates the transcription of PhGI2 by yet unknown mechanisms, which makes a separation of paralog function difficult.
Expression patterns of the core clock genes LHY and TOC1 were similar in wildtype and silenced lines and similar to Arabidopsis, where LHY peaks late during the night and is lowest at the onset of the night while TOC1 expression is counterphased to LHY, forming a negative feedback loop5. This feedback control system therefore was not altered by PhGI1 silencing. Different from Arabidopsis, were ZTL messenger RNA is constitutively expressed, we observed a peak expression towards midday of PhCHL both in wildtype and silenced lines. A lack of change in PhCHL expression in silenced lines can be explained by the fact that in Arabidopsis, interaction of GI with ZTL occurs at the protein level, consisting in the facilitation of maturation of ZTL into a functional protein. ZTL targets TOC1 for proteasomal degradation56, suggesting that changes of TOC1 protein would also be expected. In Arabidopsis, GI and ELF4 have a synergistic effect on endogenous clock regulation, showing epistatic interactions38. Furthermore, GI and ELF4 proteins interact physically to form discrete nuclear bodies57 but no direct interaction on the expression level is reported, which might explain the similarity in expression pattern between PhGI1 silenced lines and wildtype.
In petal tissue, silencing of PhLHY resulted in a phase-advance of GI peak expression of 4 hours58, indicating that disturbance of the normal expression pattern of clock genes may alter the rhythmicity of the same and other clock genes. Similarly, the silencing of GI1 here led to a significant prolongation of rhythmic period of 3 hours for the genes PhGI1 and PhCHL in all silenced lines.
Changes in the vegetative growth of PhGI1 transgenic lines was characterized by an increased leaf size of basal and apical leaves, and an augmentation in the basal internode length. However, medium and apical internodes were shorter, thus compensating in total plant length, which was not altered. It is well reported that nitrogen concentration diminishes with increasing shoot biomass during plant growth as result of N dilution59 and this dilution effect might have contributed to the progressive reduction in internode length in PhGI1 transgenic lines. Next to the changes in internode length and leaf size, we also observed a structural change in growth characterized by an increased number of axillary meristems as well as a higher chlorophyll level in apical leaves. All changes together led to a bushier phenotype with darker color. In Arabidopsis, GI controls the growth of the hypocotyl38 and the loss of function of GI results, apart from late flowering, in long petioles, tall plant height and many rosette leaves60. The findings in Arabidopsis confirm the effect of GI on vegetative growth observed in the silenced Petunia lines. GIGANTEA is known as a key regulator of flowering time. In Arabidopsis, GI mutation leads to a late-flowering phenotype in LD conditions30. The role of GI in flowering is conferred through its control over CO and FT mRNA expression levels under inductive conditions as found in different plant species30. A second pathway involving GI is CONSTANS (CO) independent and involves GI regulation of miR172, which than controls FT induction and flowering32 We did not observe a switch in inflorescence phase of iRNA::PhGI1 lines compared to wildtype lines, indicating that PhGI1 does not share a function in controlling flowering time with AtGI. Future research will show whether a case of subfunctionalization has occurred in Petunia were only the second copy of GI in petunia, PhGI2, affects flowering time. However, the relation between late flowering time and increased biomass seen in Arabidopsis is broken in Petunia as the PhGI1 silenced lines did not flower later than wild type. In fact, mutant combinations of RVE genes in Arabidopsis also disrupt the correlation between biomass production and flowering via changes in PIF gene expression61. Altogether, our results indicate that PhGI1 has an effect on plant growth coordination. Interestingly, the analyzed parameters on vegetative and generative growth in general showed a stronger reduction in T1 lines that T2 lines as compared to the wild type. This might be due to the exposure to lower night temperatures of greenhouse grown T2 lines compared to the growth chamber grown T1 lines, as it is known that siRNA generation and silencing is inhibited by low temperatures62.
PhGI1 silenced lines were characterized by a reduction in the number of flower buds, the appearance of two flower buds at the bifurcation point of the inflorescence meristem, of which one flower bud aborted, and an increased overall incidence of premature failure in floral development. All these phenomena were not described until now for any other GI mutant. The appearance of ectopic flower buds in the bifurcation point of the inflorescence meristem was not described for any other mutant in Petunia hybrida. Mutants affecting flower bud appearance described so far are double top (dot) and aberrant flower (alf), characterized by a failure to develop flowers and extra petals (exp) and evergreen (evg), were the inflorescence forms a solitary flower63,64. Aborted flowers clearly show carpel and stamen tissues indicating that flower abortion occurred following the activation of genes specifying floral organ identity. On the other hand, the overall reduced number of flower buds suggests an effect of PhGI1 silencing on upstream events, possibly related the flower-meristem-identity genes PETUNIA FLOWERING GENE (PFG) and ALF (ABERRANT LEAF AND FLOWER)41,63. Mutants showing a developmental arrest in flower bud development all belong to the group of gibberellin deficient mutants, including gibberellin deficient (ga-2)65 and gib-166 from tomato or ga1-1 from Arabidopsis67. The promotion of petal, stamen and anther development in Arabidopsis was proposed to occur by opposing the action of the DELLA proteins RGA, RGL1 and RGL268. As mentioned above, GI is a negative regulator of growth, as GI loss of function mutants show taller plant height60 and longer hypocotyls. However, the function of GI in flower development seems inverse, as flowers either aborted or showed a reduction in corolla and tube size. The reduced size was accompanied by a significant reduction in cell size, indicating that flower size changes are, at least in part, due to a reduced cell expansion, even so we cannot rule out a possible effect over cell division. Growth of lateral organs starts with cell division, followed by cell expansion during later stages of development69,70,71,72. Our results indicate that PhGI1 function on lateral organ growth depends on the acquired meristem identity and that the growth promoting function of PhGI1 during flower development is restricted rather to developmental stages following organ differentiation, when growth relies on cell expansion.
The floral fragrance in Petunia hybrida is dominated by volatile benzenoids, which mostly derive from trans-cinnamic acid, whose precursor is phenylalanine. The production of phenylalanine is controlled by ODORANT1 (ODO1), a key volatile regulator and member of the R2R3-type MYB family, which controls the synthesis of precursors of the shikimate pathway73. The main volatile, methyl benzoate, has its maximum emission at night74. It is produced from benzoic acid, whose synthesis might be controlled by PAL75. Wildtype Petunia observed here showed a rhythmic emission pattern with maximal emission during the night and methyl benzoate continuously was the major compound throughout the day. Differences between the wildtype and the silenced lines consisted (1) in a lower emission level, (2) in slight changes in the relative abundance of the trans-cinnamic acid derivatives benzyl alcohol, ethyl benzoate and benzyl benzoate and (3) a mayor contribution of isoeugenol to the volatile profile in the morning. Isoeugenol also derives from phenylalanine, but its direct precursor was suggested to be ferrulic acid, produced from trans-cinnamic acid through coumaric acid and caffeic acid. Our finding suggests that GI interacts in the rhythmic fine tuning of volatile biosynthesis and the daily emission profile of volatiles derived through the phenylalanine pathway. We cannot exclude that some changes in emission quantity and quality shortly after sampling might be related to wounding, as it was shown that stress conditions and membrane damage may affect VOC generation76.
While the plant circadian clock coordinates environmental inputs into basic processes such as primary and secondary metabolism, cell division or cell expansion, in this work we uncover undescribed functions of PhGI1 on overall inflorescence architecture. It remains to be determined if the phenotypes found in this study are directly controlled by the clock or are specific functions resulting from neofunctionalization of GI genes in Petunia.
Methods
Plant material, growth conditions and sampling
Wild type Petunia hybrida plants of the double haploid variety ‘Mitchell W115’ as well as silenced lines of the T1 generation of PhGI1 and their non-transgenic siblings were cultured using a commercial substrate (Universal Substrate, Floragard Betriebs GmbH, Oldenburg, Germany) in a growth chamber under conditions of 16 hours light/ 8 hours darkness, a light intensity of 250 μE m−2 s−1, and a constant temperature of 26 ± 1 °C. A T2 generation of PhGI1 was grown in a greenhouse under natural long-day conditions. Plants were watered as required and transplanted to fresh substrate twice during the growth phase.
Phenotyping of vegetative and generative traits, including the size of three leaves and flowers, internode length, flower number, flowering time and relative chlorophyll content was performed. Parameters were evaluated from three wild type plants and 2–3 plants of each iRNA::PhGI1 line. Of each autopollinated T1 plant, T2 plants were propagated, of which were characterized at least three plants per silenced line, in order to confirm the RNA interference associated phenotypes.
For PhGI1 expression analysis, as well as other circadian rhythm related genes, three samples of young leaves, from each of the three independent iRNA::PhGI1 transgenic lines as well as wildtype plants, were sampled under the aforementioned growth chamber conditions. Tissue sampling was performed every three hours. For the analysis of the expression under continuous darkness, plants were initially acclimated during 4–5 days to conditions of 16 hours of light / 8 hours of darkness, after which we proceeded to keep the plants in continuous darkness for 24 consecutive hours. The collected tissues were immediately frozen in liquid nitrogen and stored at −80 °C until further analysis. To measure the progress of time in hours, we used the ZEITGEBER time scale. The term ZEITGEBER, from the German “time giver”, is often used to indicate an external environmental factor capable of synchronizing the biological clock of an organism. We considered ZEITGEBER 0 (ZT0) as the time lights were turned on.
For the analysis of VOC profiles, we sampled three flowers per plant at 2–3 days after flower opening at ZT0. The measurement of VOC emission was performed as described previously77,78. Briefly, flowers were placed in a glass beaker with a solution of 4% of glucose inside a desiccator and emitted volatiles were collected using the SPME methods from the headspace during 24 hours as well as every three hours during 24 hours under 16 h/8 h photoperiod, followed by GC/MS. Volatiles were expressed as integrated peak area divided by flower fresh weight79. VOCs that contributed with at least 2% to the total emission are considered as main VOCs.
Silencing of PhGI1: Generation of vector constructs and transformation
For vector construction, we selected a fragment of the 3′ untranslated region of PhGI1 that would discriminate between PhGI1 and PhGI2. The sequence information for the comparison between PhGI1 and PhGI2 was obtained from the genomic clones PhGI1 (Peaxi132Scf1428Ctg026) and PhGI2 (Peaxi132Scf1428Ctg060) identified in P. hybrida W115 (Fig. S1). Based on this comparison, we selected a DNA fragment of 225 bp from PhGI1, that showed maximal sequence difference with PhGI2 (Fig. S2) and this fragment was PCR-amplified using site-specific primers containing the attB1 and attB2 sites for Gateway recombination80. Genomic DNA was used as template for all fragment amplification. Each fragment was first recombined into the entry vector pDONR201 (Invitrogen) and then recombined into the final destination vector pHELLSGATE12 in order to obtain hairpin-like structures. All primers used for plasmids generation are listed in Table S5.
The W115 Mitchell double haploid was transformed as described before81 using Agrobacterium tumefaciens strain EHA105. Shoots, developed under selective conditions, were confirmed as transformed through PCR detection of the selection marker gene nptII (T0, T1) and DNA blot analysis (T0) with a nptII DIG-labeled DNA probe (Fig. S4)82.
Circadian gene expression analysis
Total RNA from leaves was isolated using a phenol:chloroform based protocol83. Following spectrophotometric quantification (NanoDrop2000), equal amounts of RNA were used to synthesize cDNA according to the manufacturer’s instructions (Maxima First Strand cDNA Synthesis Kit for RT-qPCR, with dsDNase Thermofisher (https://www.thermofischer.com/, catalog number: K1641).
The gene ACTIN 11 (ACT), previously selected as valuable housekeeping gene for Petunia leaves and petals under circadian conditions47 was used as reference gene for relative expression quantification of clock genes. Primers for PhGI1, PhGI2 and other clock genes (Table S5) were designed using pcrEfficiency software84. Quantitative PCR and melting point analysis were performed as described previously22. Three biological and two technical replicas were analyzed for each sample.
Chlorophyll content
Chlorophyll content was determined in basal, medium and apical leaves of wildtype plants and three silenced lines of PhGI1 from T1 generation kept under 16:8 LD light regime. Relative chlorophyll content was calculated using a CM-500 Chlorophyll Meter (SolfrancTecnologías SL) based on measuring light penetration coefficient in a two wavelength range corresponding to red light and IR light.
Scanning electron microscopy analysis
We observed petal cell size in the corolla and the floral tube of silenced lines from T1 generation and non-transgenic siblings. The two areas were separated with a scalpel blade. From the corolla, we prepared two zones for further analysis of cell size, the distal outer zone and a proximal zone near the tube. Petal sections had a size of approximately 0.75 cm2. Cell size was calculated measuring the area of 50 cells from 3 different flowers of 3 plants by using the program ImageJ (ttps://imagej.nih.gov/ij/download.html).
The floral meristems were sampled from flowers of GI1 silenced line from both positions, those that develop into mature flowers and those that develop into aborted flowers. Preparation of flower buds for scanning electron microscopy consisted in the removal of the sepals. All tissues were dehydrated as previously described85, followed by critical point drying.
Data analysis procedures
Expression of circadian genes relative to the reference genes was analyzed applying the comparative CT method86,87 as well as using group-wise comparison with the REST Program88. The JTK-Cycle algorithm from the MetaCycle R package (R version 3.3.2)88,89 was applied in order to detect rhythmicity in gene expression. Significance differences among data were determined based on Fisher´s F-test and Student´s T-Test after testing data for non-normal distributions.
Significance statement
The Gigantea gene appeared in land plants and is considered as an activator of floral transition. In Petunia it has several functions including repression of vegetative biomass accumulation, and ectopic flower initiation. In PhGI1-silenced plants, flowers either aborted or grew to small sizes, emitting low quantities of scent. PhGI1 thus shows a new set of functions during flower development.
References
Staiger, D., Shin, J., Johansson, M. & Davis, S. J. The circadian clock goes genomic. Genome Biol. 14, 208 (2013).
Pokhilko, A. et al. The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops. Mol. Syst. Biol. 8, 574 (2012).
Mas, P. Circadian clock signaling in Arabidopsis thaliana: from gene expression to physiology and development. Int. J. Dev. Biol. 49, 491–500 (2005).
Kikis, E. A., Khanna, R. & Quail, P. H. ELF4 is a phytochrome-regulated component of a negative-feedback loop involving the central oscillator components CCA1 and LHY. Plant J. 44, 300–313 (2005).
Gendron, J. M. et al. Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc. Natl. Acad. Sci. USA 109, 3167–72 (2012).
Adams, S., Manfield, I., Stockley, P. & Carré, I. A. Revised Morning Loops of the Arabidopsis Circadian Clock Based on Analyses of Direct Regulatory Interactions. PLoS ONE 10 (2015).
Más, P., Kim, W.-Y., Somers, D. E. & Kay, S. A. Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426, 567–570 (2003).
Cha, J.-Y. et al. GIGANTEA is a co-chaperone which facilitates maturation of ZEITLUPE in the Arabidopsis circadian clock. Nat. Commun. 8, 3 (2017).
Kim, W.-Y. et al. ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 449, 356–360 (2007).
Bancos, S. et al. Diurnal Regulation of the Brassinosteroid-Biosynthetic CPD Gene in Arabidopsis. Plant Physiol. 141, 299–309 (2006).
Nozue, K. et al. Rhythmic growth explained by coincidence between internal and external cues. Nature 448, 358–361 (2007).
Thines, B. & Harmon, F. G. Four easy pieces: mechanisms underlying circadian regulation of growth and development. Curr. Opin. Plant Biol. 14, 31–7 (2011).
Soy, J. et al. Molecular convergence of clock and photosensory pathways through PIF3–TOC1 interaction and co-occupancy of target promoters. Proc. Natl. Acad. Sci. 201603745, https://doi.org/10.1073/pnas.1603745113 (2016).
Fung-Uceda, J. et al. The Circadian Clock Sets the Time of DNA Replication Licensing to Regulate Growth in Arabidopsis. Dev. Cell 45, 101–113.e4 (2018).
Farré, E. M. & Weise, S. E. The interactions between the circadian clock and primary metabolism. Curr. Opin. Plant Biol. 15, 293–300 (2012).
Marcolino-Gomes, J. et al. Diurnal oscillations of soybean circadian clock and drought responsive genes. PloS One 9, e86402 (2014).
Weiss, J. et al. Diel pattern of circadian clock and storage protein gene expression in leaves and during seed filling in cowpea (Vigna unguiculata). BMC Plant Biol. 18, 33–53 (2018).
Ni, Z. et al. Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature 457, 327–331 (2009).
Atamian, H. S. et al. Circadian regulation of sunflower heliotropism, floral orientation, and pollinator visits. Science 353, 587–590 (2016).
Edwards, K. D. et al. Circadian clock components control daily growth activities by modulating cytokinin levels and cell division-associated gene expression in Populus trees: Control of growth in Populus. Plant Cell Environ. 41, 1468–1482 (2018).
Yon, F. et al. Silencing Nicotiana attenuata LHY and ZTL alters circadian rhythms in flowers. New Phytol. 209, 1058–1066 (2015).
Terry, M. I. et al. The Petunia CHANEL Gene is a ZEITLUPE Ortholog Coordinating Growth and Scent Profiles. Cells 8, 343 (2019).
Terry, M. I., Pérez-Sanz, F., Navarro, P. J., Weiss, J. & Egea-Cortines, M. The Snapdragon LATE ELONGATED HYPOCOTYL Plays A Dual Role in Activating Floral Growth and Scent Emission. Cells 8, 920 (2019).
Fenske, M. P. et al. Circadian clock gene LATE ELONGATED HYPOCOTYL directly regulates the timing of floral scent emission in Petunia. Proc. Natl. Acad. Sci., https://doi.org/10.1073/pnas.1422875112 201422875–201422875 (2015).
Corellou, F. et al. Clocks in the Green Lineage: Comparative Functional Analysis of the Circadian Architecture of the Picoeukaryote Ostreococcus. Plant Cell 21, 3436–3449 (2009).
Morant, P. E. et al. A robust two-gene oscillator at the core of Ostreococcus tauri circadian clock. Chaos 20 (2010).
Djouani-Tahri, E. B. et al. A eukaryotic LOV-histidine kinase with circadian clock function in the picoalga Ostreococcus. Plant J. 65, 578–588 (2011).
Linde, A. et al. Early evolution of the land plant circadian clock. New Phytol., https://doi.org/10.1111/nph.14487 (2017).
Kubota, A. et al. Co-option of a photoperiodic growth-phase transition system during land plant evolution. Nat. Commun. 5, 3668 (2014).
Fowler, S. GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 18, 4679–4688 (1999).
Mizoguchi, T. et al. Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis. Plant Cell 17, 2255–70 (2005).
Jung, J.-H. et al. The GIGANTEA -Regulated MicroRNA172 Mediates Photoperiodic Flowering Independent of CONSTANS in Arabidopsis. Plant Cell 19, 2736–2748 (2007).
Sawa, M. & Kay, S. A. GIGANTEA directly activates Flowering Locus T in Arabidopsis thaliana. Proc. Natl. Acad. Sci. 108, 11698–11703 (2011).
Ding, J. et al. GIGANTEA- like genes control seasonal growth cessation in. Populus. New Phytol. 218, 1491–1503 (2018).
Dalchau, N. et al. The circadian oscillator gene GIGANTEA mediates a long-term response of the Arabidopsis thaliana circadian clock to sucrose. Proc. Natl. Acad. Sci. USA 108, 5104–9 (2011).
Park, H. J., Kim, W.-Y. & Yun, D.-J. A role for GIGANTEA: keeping the balance between flowering and salinity stress tolerance. Plant Signal. Behav. 8, e24820 (2013).
Cao, S., Ye, M. & Jiang, S. Involvement of GIGANTEA gene in the regulation of the cold stress response in Arabidopsis. Plant Cell Rep. 24, 683–690 (2005).
Kim, Y. et al. GIGANTEA and EARLY FLOWERING 4 in Arabidopsis Exhibit Differential Phase-Specific Genetic Influences over a Diurnal Cycle. Mol. Plant 5, 678–687 (2012).
Tseng, T.-S. SPINDLY and GIGANTEA Interact and Act in Arabidopsis thaliana Pathways Involved in Light Responses, Flowering, and Rhythms in Cotyledon Movements. PLANT CELL ONLINE 16, 1550–1563 (2004).
Souer, E. et al. Genetic control of branching pattern and floral identity during Petunia inflorescence development. Development 125, 733–742 (1998).
Immink, R. G. et al. A petunia MADS box gene involved in the transition from vegetative to reproductive development. Dev. Camb. Engl. 126, 5117–5126 (1999).
Souer, E. et al. Patterning of inflorescences and flowers by the F-Box protein DOUBLE TOP and the LEAFY homolog ABERRANT LEAF AND FLOWER of petunia. Plant Cell 20, 2033–2048 (2008).
Eckardt, N. A. DOT/UFO Emerges as a Key Factor in Inflorescence Patterning. Plant Cell 20, 2003–2005 (2008).
Rebocho, A. B. et al. Role of EVERGREEN in the development of the cymose petunia inflorescence. Dev. Cell 15, 437–447 (2008).
Rijpkema, A. S., Vandenbussche, M., Koes, R., Heijmans, K. & Gerats, T. Variations on a theme: Changes in the floral ABCs in angiosperms. Semin. Cell Dev. Biol. 21, 100–107 (2010).
Bombarely, A. et al. Insight into the evolution of the Solanaceae from the parental genomes of Petunia hybrida. Nat. Plants 2, 16074 (2016).
Terry, M. I., Carrera-Alesina, M., Weiss, J. & Egea-Cortines, M. Molecular and transcriptional structure of the petal and leaf circadian clock in Petunia hybrida., http://biorxiv.org/lookup/doi/10.1101/641639, https://doi.org/10.1101/641639 (2019).
Somers, W. S., Tang, J., Shaw, G. D. & Camphausen, R. T. Insights into the Molecular Basis of Leukocyte Tethering and Rolling Revealed by Structures of P- and E-Selectin Bound to SLeX and PSGL-. 13 (2000).
Yon, F. et al. Identification and characterization of circadian clock genes in a native tobacco, Nicotiana attenuata. BMC Plant Biol. 12, 172 (2012).
de Montaigu, A., Tóth, R. & Coupland, G. Plant development goes like clockwork. Trends Genet. 26, 296–306 (2010).
Más, P., Alabadí, D., Yanovsky, M. J., Oyama, T. & Kay, S. A. Dual Role of TOC1 in the Control of Circadian and Photomorphogenic Responses in Arabidopsis. Plant Cell 15, 223–236 (2003).
Lonardi, S. et al. The genome of cowpea (Vigna unguiculata [L.] Walp.). Plant J. 98, 767–782 (2019).
Lu, S. X. et al. CCA1 and ELF3 Interact in the control of hypocotyl length and flowering time in Arabidopsis. Plant Physiol. 158, 1079–88 (2012).
Park, D. H. et al. Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 285, 1579–82 (1999).
Jackson, A. L. et al. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 21, 635–637 (2003).
Ito, S., Song, Y. H. & Imaizumi, T. LOV domain-containing F-box proteins: light-dependent protein degradation modules in Arabidopsis. Mol. Plant 5, 573–82 (2012).
Kim, Y. et al. ELF4 Regulates GIGANTEA Chromatin Access through Subnuclear Sequestration. Cell Rep. 3, 671–677 (2013).
Fenske, M. P. & Imaizumi, T. Circadian Rhythms in Floral Scent Emission. Front. Plant Sci. 7, 462 (2016).
Bélanger, G. & Gastal, F. Nitrogen utilization by forage grasses. Can. J. Plant Sci. 80, 11–20 (2000).
Hwang, C., Park, J., Lee, B. & Cheong, H. Loss of Function in GIGANTEA Gene is Involved in Brassinosteroid Signaling. J. Chosun Nat. Sci. 4, 113–120 (2011).
Gray, J. A., Shalit-Kaneh, A., Chu, D. N., Hsu, P. Y. & Harmer, S. L. The REVEILLE Clock Genes Inhibit Growth of Juvenile and Adult Plants by Control of Cell Size. Plant Physiol. 173, 2308–2322 (2017).
Szittya, G. et al. Low temperature inhibits RNA silencing-mediated defence by the control of siRNA generation. EMBO J. 22, 633–640 (2003).
Souer, E. et al. Patterning of Inflorescences and Flowers by the F-Box Protein DOUBLE TOP and the LEAFY Homolog ABERRANT LEAF AND FLOWER of Petunia. Plant Cell 20, 2033–2048 (2008).
Rebocho, A. B. et al. Role of EVERGREEN in the Development of the Cymose Petunia Inflorescence. Dev. Cell 15, 437–447 (2008).
Nester, J. E. & Zeevaart, J. A. D. FLOWER DEVELOPMENT IN NORMAL TOMATO AND A GIBBERELLIN-DEFICIENT (ga-2) MUTANT. Am. J. Bot. 75, 45–55 (1988).
Jacobsen, S. E. & Olszewski, N. E. Characterization of the Arrest in Anther Development Associated with Gibberellin Deficiency of the gib-1 Mutant of Tomato. Plant Physiol. 97, 409–414 (1991).
Goto, N. & Pharis, R. P. Role of gibberellins in the development of floral organs of the gibberellin-deficient mutant, ga1-1, of Arabidopsis thaliana. Can. J. Bot. 77, 944–954 (1999).
Cheng, H. et al. Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development 131, 1055–1064 (2004).
Reale, L. et al. Patterns of cell division and expansion in developing petals of Petunia hybrida. Sex. Plant Reprod. 15, 123–132 (2002).
Laitinen, R. A. E. et al. Analysis of the floral transcriptome uncovers new regulators of organ determination and gene families related to flower organ differentiation in Gerbera hybrida (Asteraceae). Genome Res. 15, 475–486 (2005).
Anastasiou, E. et al. Control of Plant Organ Size by KLUH/CYP78A5-Dependent Intercellular Signaling. Dev. Cell 13, 843–856 (2007).
Kazama, T., Ichihashi, Y., Murata, S. & Tsukaya, H. The Mechanism of Cell Cycle Arrest Front Progression Explained by a KLUH/CYP78A5-dependent Mobile Growth Factor in Developing Leaves of Arabidopsis thaliana. Plant Cell Physiol. 51, 1046–1054 (2010).
Verdonk, J. C., Haring, M. A., van Tunen, A. J. & Schuurink, R. C. ODORANT1 regulates fragrance biosynthesis in petunia flowers. Plant Cell 17, 1612–24 (2005).
Boatright, J. et al. Understanding in vivo benzenoid metabolism in petunia petal tissue. Plant Physiol. 135, 1993–2011 (2004).
Kolosova, N., Sherman, D., Karlson, D. & Dudareva, N. Cellular and subcellular localization of S-adenosyl-L-methionine:benzoic acid carboxyl methyltransferase, the enzyme responsible for biosynthesis of the volatile ester methylbenzoate in snapdragon flowers. Plant Physiol. 126, 956–64 (2001).
Loreto, F., Barta, C., Brilli, F. & Nogues, I. On the induction of volatile organic compound emissions by plants as consequence of wounding or fluctuations of light and temperature. Plant Cell Environ. 29, 1820–1828 (2006).
Manchado-Rojo, M., Delgado-Benarroch, L., Roca, M. J., Weiss, J. & Egea-Cortines, M. Quantitative levels of Deficiens and Globosa during late petal development show a complex transcriptional network topology of B function. Plant J. Cell Mol. Biol. 72, 294–307 (2012).
Ruíz-Ramón, F., Águila, D. J., Egea-Cortines, M. & Weiss, J. Optimization of fragrance extraction: Daytime and flower age affect scent emission in simple and double narcissi. Ind. Crops Prod. 52 (2014).
Ruiz-Hernández, V., Roca, M. J., Egea-Cortines, M. & Weiss, J. A comparison of semi-quantitative methods suitable for establishing volatile profiles. Plant Methods 14, 67 (2018).
Helliwell, C. & Waterhouse, P. Constructs and methods for high-throughput gene silencing in plants. Methods San Diego Calif 30, 289–95 (2003).
Manchado-Rojo, M., Weiss, J. & Egea-Cortines, M. Validation of Aintegumenta as a gene to modify floral size in ornamental plants. Plant Biotechnol. J. 12 (2014).
Southern, E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98, 503–517 (1975).
Box, M. S., Coustham, V., Dean, C. & Mylne, J. S. Protocol: A simple phenol-based method for 96-well extraction of high quality RNA from Arabidopsis. Plant Methods 7, 7 (2011).
Mallona, I., Weiss, J. & Egea-Cortines, M. pcrEfficiency: a Web tool for PCR amplification efficiency prediction. BMC Bioinformatics 12, 404 (2011).
Delgado-Benarroch, L., Causier, B., Weiss, J. & Egea-Cortines, M. FORMOSA controls cell division and expansion during floral development in Antirrhinum majus. Planta 229 (2009).
Schmittgen, T. D. & Livak, K. J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108 (2008).
Pfaffl, M. W., Horgan, G. W. & Dempfle, L. Relative expression software tool (REST(C)) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucl Acids Res 30, e36- (2002).
Hughes, M. E., Hogenesch, J. B. & Kornacker, K. JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J. Biol. Rhythms 25, 372–380 (2010).
Wu, G., Anafi, R. C., Hughes, M. E., Kornacker, K. & Hogenesch, J. B. MetaCycle: an integrated R package to evaluate periodicity in large scale data. Bioinformatics 32, 3351–3353 (2016).
Acknowledgements
We would like to acknowledge María José Roca and Julia Muñoz for technical assistance. This research was funded by Fundación Seneca 19398/PI/14, 19895/GERM/15 and MC BFU-2017 88300-C2-1-R and BFU-2017 88300-C2-2-R.
Author information
Authors and Affiliations
Contributions
Conceived and coordinated the study, J.W., C.P. and M.E.C.; Research methodology, C.B., C.P., M.E.C. and J.W.; Formal analysis, C.B., C.P., M.E.C. and J.W.; Investigation, C.B., C.P., M.E.C. and J.W.; Resources, J.W., C.P. and M.E.C.; Data curation, C.B., M.E.C. and J.W.; Writing—original draft preparation, C.B., M.E.C. and J.W.; Writing—review and editing, C.B., C.P., M.E.C. and J.W.; Supervision, J.W., C.P. and M.E.C.; Project administration, J.W., C.P. and M.E.C.; Funding acquisition, J.W., C.P. and M.E.C.; All authors revised and approved the final draft.
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Brandoli, C., Petri, C., Egea-Cortines, M. et al. The clock gene Gigantea 1 from Petunia hybrida coordinates vegetative growth and inflorescence architecture. Sci Rep 10, 275 (2020). https://doi.org/10.1038/s41598-019-57145-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-57145-9
This article is cited by
-
Effects of the repression of GIGANTEA gene StGI.04 on the potato leaf transcriptome and the anthocyanin content of tuber skin
BMC Plant Biology (2022)
-
Artificial polyploidy induction for improvement of ornamental and medicinal plants
Plant Cell, Tissue and Organ Culture (PCTOC) (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.