Abstract
We developed a test to predict which patients will achieve pathological complete response (pCR) to neoadjuvant chemotherapy (NAC) and which will have residual disease (RD). Gene expression data from pretreatment biopsies of patients with all breast cancer subtypes were combined into a 519-patient cohort containing 177 TNBC patients. Two RNA classifiers of 16 genes each were sequentially applied to the total cohort, classifying patients into 3 distinct classes. The test performance was further validated in an independent 304-patient cohort. The test accurately identified 70.5% (79/112) of pCR and 83.5% (340/407) of RD patients in the total population, and 75.0% (45/60) of pCR and 75.2% (88/117) of RD patients in the TNBC subset. For the independent cohort, the test identified 91.5% RD patients in the total population and 86.2% RD patients in the TNBC subset. However, the identification of pCR in both total and TNBC population are as low as 21.1% and 30%, respectively. The TNBC RD patients were subdivided by our classifiers, with one class showing significantly higher levels of Ki67 expression and having significantly poorer survival rates than the other classes. This stratification of patients may allow predicted residual disease classes to be assigned an alternative therapy.
Similar content being viewed by others
Introduction
Triple-negative breast cancer (TNBC), characterized by lack of expression of the estrogen (ER), progesterone (PgR), and erb-b2 receptor tyrosine kinase 2 (HER2) receptors, is a particularly problematic form of breast cancer due to aggressive growth, high recurrence rates and poor long-term survival1,2. TNBC represents 15–20% of newly diagnosed breast cancers in the United States3. Achieving pathological complete response (pCR) to neoadjuvant chemotherapy (NAC) is a surrogate marker and predictor of long-term outcomes, especially for TNBC4,5,6,7,8,9,10,11. Thus, NAC can allow for an early evaluation of the effectiveness of systemic therapy. Since pCR is correlated with prediction of 5-year disease free survival, biomarker development in this setting can establish efficacy which can then be utilized in both the neoadjuvant and adjuvant settings. In addition, only about 20% of breast cancer patients achieve pCR11, causing unnecessary morbidity for the other 80% receiving high-toxicity treatment with limited benefit. Predicting which patients will have pCR or residual disease (RD) provides physicians with an opportunity to improve treatment planning with more aggressive or novel treatments, while preventing overtreatment in populations expected to achieve pCR with the standard of care.
pCR predictors have been proposed, but they either did not achieve the necessary levels of positive and negative predictive values for clinical utility, suffered from small sample sizes, lacked validation data, or were not applicable to TNBC12,13,14,15,16,17,18,19,20,21. While effective molecular tests exist to guide treatment for estrogen receptor (ER) or HER2+ tumors, there are no tests in clinical use to stratify TNBC. High levels of tumor infiltrating lymphocytes (TILs) have shown a correlation with increased pCR rates in TNBC, in that 31% of the low-TIL patients achieved pCR compared to 50% of the high-TIL patients22. But this correlation is not strong enough to use as a predictive model, because it would predict pCR for all high-TIL tumors and would be wrong for 50% of the patients. Patients with grade 3 tumors, containing cells morphologically different from healthy cells, were found to have higher pCR rates in platinum-based neoadjuvant therapy than patients with lower-grade tumors23. However, the reported odds ratio of this finding was 1.73, which converts to a probability of 63.4%, and this would also be its accuracy if used as a predictive test. Gene expression profiling has been used to define four TNBC subtypes with significant differences in their pCR rates15. But again, even the subtype most associated with pCR contained only 41% of the patients that achieved pCR, which is still not a clinically useful predictor. There is a strong unmet need for a predictor of response to neoadjuvant chemotherapy for TNBC patients that is sufficiently accurate to use in treatment decisions.
We developed a test called BA100 to predict which patients are likely to achieve pCR or RD to standard NAC using gene expression profiling of 325 novel biomarkers that are associated with non-malignant breast epithelial cell organization and that correlate with breast cancer clinical outcomes24,25,26, 23 TNBC-related genes27, and a unique and proprietary machine learning algorithm to select and rank informative genes. While the 325 genes are mainly involved in pathways such as proliferation, DNA repair, cell survival, metabolisms, cell migration and adhesion, two genes in the 23 TNB-related genes are likely involved in cell immunity. One gene is HLA-DPA1, which encodes a major histocompatibility complex, class II, DP alpha 1 protein. This protein is mainly expressed in antigen presenting cells (APC) including macrophages, dendritic cells, as well as B lymphocytes as hetero-dimers with HLA-DPB, and plays an essential role in human immune system by presenting the extracellular peptides to T cells28. The other gene VTCN1, encoding V-set domain containing T cell activation inhibitor 1, belongs to the B7 costimulatory protein family. Proteins in this family are also present mostly on APCs, and interact with ligands that bind to receptors on cytotoxic T cells. VTCN1 was found to negatively regulate cell immune response, and expression of VTCN1 negatively correlates with patient’s clinical outcome29. The results show that in addition to stratifying patients into pCR and RD classes, the BA100 test also identifies a third class of RD patients with worse survival than the others. We characterized these three classes of patients to see if some are associated with higher residual cancer burden (RCB), which is clinically similar to pCR/RD classification; whether they correspond to some of the breast cancer subtypes defined by the PAM50 molecular classifier; and whether they differ in Ki-67 or androgen receptor (AR) expression, which are known to impact the aggressiveness of breast cancers. We also compare our BA100 predictor to a previously published one called DLDA3021,30.
Methods
Patients and data sets
A patient data set of Affymetrix Human Genome U133A GeneChip® Array gene expression with associated outcomes and treatment data was constructed from three patient cohorts with stage II-III invasive breast cancer treated with standard NAC incorporating a taxane, an anthracycline, and cyclophosphamide (AC-T), or additionally 5-fluorouracil (T-FAC). The I-SPY1 (Investigation of Serial Studies to Predict Your Therapeutic Response with Imaging and Molecular Analysis) trial5 contained breast cancer patients of all subtypes who received AC-T NAC, with HER2+ patients also receiving trastuzumab, while two studies from NCBI’s Gene Expression Omnibus (GEO), GSE25055 and GSE2506513, contained patients receiving T-FAC. After duplicates and samples with missing data were removed, a combined cohort of 519 patients remained. The cohort included 56.1% of patients with ER+/HER2- and 34.1% with TNBC, based on the annotated hormone receptors and HER2 status provided in the studies used. Suppl. Table 1 shows TNBC patient demographics and histopathological information. In short, 85.3% of TNBC patients were grade 3, with 33.9% achieving pCR, and 66.1% had residual disease.
An independent external validation set of 304 patients was constructed from GEO studies GSE2019431, GSE2027121, and GSE3264632 by removing duplicate patient entries between each other and the 519-patient training and testing cohort. Patient demographics are shown in Supplemental Table 2. The pCR rate for the 304-patient external validation cohort was 18% for all patients and 24% in TNBC.
Expression data preparation
Affymetrix’s CEL files for the 519-patient dataset were downloaded from GEO, imported into R using the BioConductor Affymetrix package, processed using the BioConductor mas5 package, subjected to batch correction using ComBat from the sva R package, and quantile normalized. CEL files for the 304-member data set were treated identically with the exception that they were normalized using quantiles obtained from 519-patient training and test set, and no batch correction was performed, in order to simulate real-world single-sample testing.
Development of RNA-biomarker classifiers
The original patient cohort was divided into a training set of 80% patients and a testing set of 20% patients. The training and testing sets had similar fractions of pCR and RD patients, and similar representations of ER, Her2, and PGR status. The training cohort was further subdivided into temporary 80%/20% subsets as part of thousands of rounds of model development, with each round having different samples selected from the pCR and RD groups. Starting with the 325 genes described above24,25,26, a proprietary algorithm based on backward regression general linear modeling (BRGLM) was used to select a smaller set of genes with the greatest predictive power. This resulted in 16 genes that were used to fit a linear regression model. To reduce the numbers of false positives detected among TNBC patients, we added genes from a TNBC signature27, repeated the modeling process using only patients that were predicted to achieve pCR by the first model, and developed a second 16-gene classifier. The use of two sequential classifiers containing different genes allowed us to combine a general predictor for all disease subtypes with a second predictor that improves the classification of TNB patients. It also made possible the identification of the third class of RD patients with worse prognosis. This modeling method has also been described in two conference proceedings. The sequential 16-gene/16-gene classifiers were applied to the 519-patient cohort and the 304-patient external validation cohort.
Classifier 1 stratified patients into the first predicted RD group (Class 2). Classifier 2 was then applied to stratify the remaining population into a predicted pCR (Class 1) and a second group of predicted RD (Class 3) (Fig. 1A). The genes comprising the classifiers are shown in Supplemental Table 3 along with their coefficients, intercepts, and threshold values above which pCR is predicted.
Description of BA100 test. (A) A diagram showing the flow of patients from the 519-member data set through the two classifiers of BA100 and resolution into 3 classes based on unique gene profiles. “N” represents the number of patients stratified at each step. (B) Output of the BA100 scores for the total population of 519 patients. The black squares represent patients achieving pCR, the red squares those with RD and the dashed lines the cutoff values above which pCR is predicted. Scores from Classifier 1 are on Y-axis and Classifier 2 on the X-axis. The Class 1 patients are those that are predicted pCR by both classifiers in the upper right quadrant, Class 2 those predicted RD by Classifier I (bottom half) and Class 3 those that Classifier 1 predicted pCR while Classifier 2 predicted RD (upper left quadrant). (C) Output of the BA100 scores for the TNBC population of 177 patients. Description as in (B).
Kaplan-Meier plots
Distant recurrence-free survival (DRFS) data was available for a maximum of 10 years in the 518-patient cohort (1 TNBC patient from the 519-member data set lacked unambiguous survival data and was excluded). Censoring was performed as indicated within the public data files. Standard Kaplan-Meier curves were generated using R packages and the statistical significance was determined by the Cox proportional hazard model with Wald and logrank test.
Comparisons of tumor characteristics
Classifications by PAM50, residual cancer burden (RCB), and a pre-defined 30-gene predictor (DLDA30)21,30 were annotated for most of the 519-patient data set and were tallied and presented as column plots. The values for androgen receptor (AR) and Ki-67 gene expression were extracted from the expression data, segregated by BA100 class and presented as box plots. The Pearson chi-squared test of independence and t-tests were used to calculate associations of RCB, PAM50 subtype, AR expression, and Ki-67 expression with BA100 classes.
Ethics approval and consent to participate
The study does not involve human subjects.
Results
Stratification of patients
Scores from the sequential application of our two classifiers (Fig. 1A) are shown as 2-dimensional scatter plots for the total population and 177 TNBC patients, respectively in Fig. 1B,C, with pCR concentrated in the upper right quadrant corresponding to the predicted positive (class 1). Conversely, the scores for RD patients are predominately scattered over the other 3 quadrants comprising classes 2 and 3.
The results of applying BA100 to stratify patients into pCR (Class 1) and RD (Classes 2 and 3 combined) in the 519-patient data set are shown in Table 1. The pCR rates of each population are shown to range from 10.3% for ER+/HER2- to 48.5% in HER2+ patients. The test correctly stratified 70.5% (79/112) of pCR and 83.5% (340/407) of RD patients in the total population, and 60.8% (45/74) of pCR and 85.4% (88/103) of RD patients in the TNBC subset. The overall accuracy was 80.7% in the total cohort and 75.1% in the TNBC subset.
The data set included values for the DLDA30 predictor for 145 of the 177 TNBC patients and the resulting test predictions are shown in Table 1 for comparison. DLDA30 correctly stratified 37.5% (45/120) of the pCR and 76.0% (19/25) of the RD patients, with an overall accuracy of 44.1%. BA100 outperforms DLDA30 on each of these measures.
BA100 stratifies TNBC patients into distinct classes of patients
Given that unique gene expression profiles from 2 independent classifiers defined biologically distinct classes of patients (BA100 Class 1–3), we examined if the classes correlated with clinical outcomes and other biomarkers.
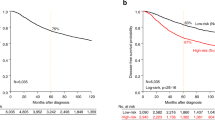
Kaplan-Meier (KM) curves with up to 10 years DRFS are shown for TNBC patients achieving pCR or RD after NAC before BA100 stratification (Fig. 2A), and with BA100’s Class 1, Class 2, and Class 3 stratification (Fig. 2B). Achieving pCR was a surrogate marker for long term survival in all cases (hazard ratio = 4.7, p = 6.5e-06). Class 3 RD patients show a statistically significant worse prognosis in comparison with Class 2 RD patients (hazard ratio = 1.88, p = 0.047) (Fig. 2C right panel) showing that BA100 stratified RD patients into two groups with different risk profiles.
KM curves showing DRFS for TNBC patients over a maximum of 10 years of follow up. (A) The total population of TNBC patients prior to BA100 stratification is divided by actual pCR (black line) and RD (red line) (pCR = 60, RD = 116). The 95% confidence intervals are indicated by the dashed lines, and the hazard ratio, p-value, and numbers of patients and scored events at various days after treatment are shown below. (B) After BA100 stratification the DRFS KM curves for each Class of TNBC patients are displayed as above. (C) The left panel shows a comparison of DRFS for the pCR patients from all three classes. The black curve is class 1, the red curve is Class 2, and the green curve is Class 3. No significant differences are noted. The right panel shows a comparison of DRFS for the RD patients in all classes with the same curve colors as in the left panel. Here, there is a significant difference seen between Class 1 pCR and Class 2 RD patients (p = 0.012) and between Class 3 and Class 2 (p = 0.047).
To test the possibility that these classifications could also predict RCB33, a measure calculated after NAC and surgery, we determined the RCB distribution across the TNBC BA100 classification based on dataset annotations (Fig. 3A). While RBC did not explain BA100 classification there were significant differences in RBC profile, with 65% of Class 1 comprised of RCB0/1, 35% and 43% of Class 2 being RCB 0/1 and RCB2 respectively, and 50% of Class 3 being RCB 3 (Pearson chi-squared p = 6.116e-06). Thus, BA100 can not only predict pCR or RD but can classify patients into classes presenting distinct RCB profiles from the initial patient biopsy, prior to any treatment.
Clinical and gene expression comparisons with TNBC classes. (A) The distributions of RCB determined from the surgical specimen after NAC are shown for each BA100 class. Blue for RCB 0/I, orange for RCB-II, and gray for RCB-III. (B) The PAM50 classifications are shown for each BA100 class. (C) The expression levels of Ki-67 are shown as a box plot for Class 1 (red), Class 2 (green), and Class 3 (blue), with each box representing the interquartile range of gene expression, and a horizontal line inside showing the median expression. The p-values of pairwise t-tests are shown, indicating that Class 2 has significantly lower Ki-67 expression than the other two classes. (D) Expression of androgen receptor in TNBC tumors for each class is shown as in (C).
We also tested if PAM50 molecular subtypes34,35 would show any association with the BA100 classes (Fig. 3B). In TNBC, the expected large basal component is consistently dominant in all of BA100 classes ranging from 66% to 88% of the populations. BA100 Classes 2 and 3 show a discrete increase in numbers of normal and HER2 molecular subtypes respectively (Pearson chi-squared p = 0.002231).
Ki-67 expression showed highly significant differences between BA100 classes. In Fig. 3C, box plots show that while TNBC classes 1 and 3 have similar Ki-67 expression, each with median expression close to 7.4 and an interquartile range of about 7.0 to 7.6, class 2 shows significantly lower levels than the other classes (Class 2 vs Class 3 p = 1.72e-7), with a median expression of 7.0 and an interquartile range of 6.8 to 7.4. Androgen receptor expression was also assessed (Fig. 3D), and Class 2 had a higher median expression of AR, but it did not meet the p < 0.05 threshold of significance. The top portion of Supplemental Table 1 shows the distribution of various TNBC tumor parameters per class. There is no significant association between TNBC classes and histologic grade, clinical stage, T-Stage, or nodal status.
Test of BA100 Against an External Validation Set
We analyzed an independent validation cohort of 304 patients treated with neoadjuvant T-FAC (demographics are shown in Supplemental Table 2) using the BA100 test. ER + /HER2- tumors comprised 44% of the cohort, TNBC tumors comprised 28%, and there was an overall pCR rate of 18.8% (Table 1 lower panel). Though the pCR rate is lower compared to the 519-member general population, we used the cohort to mimic real-life testing of sequential samples analyzed on a sample-by-sample basis without batch correction, as would be the case for use of BA100 in a clinical setting. BA100 correctly stratified 21.1% (12/57) of the pCR and 91.5% (226/247) of the RD patients in this cohort, for an overall accuracy of 78.3%. For the small TNBC subset, BA100 correctly stratified 30.0% (6/20) of the pCR and 86.2% (56/65) RD patients, for an overall accuracy of 72.9%. The prediction of RD patients in the validation cohorts had slightly better accuracy than that of the original cohort (91.5% vs 83.5%), but the accuracy of pCR predictions was much lower in the validation set (70.5% in training, 21.1% in validation). We think that the lack of batch correction in combining the three external validation data sets and the different composition of pCR vs RD, as well as Her2 patients may be contributors to the lower accuracy observed. Additional studies will be needed to further validate the BA100 test.
Discussion
Achieving pCR is associated with significantly improved DRFS in TNBC patients, and previous work also supports the importance of achieving pCR for a variety of survival metrics, suggesting that pCR should be a major goal of NAC10,33. The benefit for TNBC patients is particularly strong as the risk functions modeled by the KM curves show that only 15% of TNBC patients with pCR are expected to suffer distant recurrences as compared to 60% of the RD patients over a 10-year span (Fig. 2A). There is also benefit in identifying those patients who will have RD with standard NAC, so the treatment can be altered to increase the chances of achieving pCR. These observations spurred us to develop a method to predict which patients are likely to experience pCR with the standard of care NAC, and alternatively which patients could be spared more aggressive chemotherapy regimens or be assigned to novel treatments in clinical trials (e.g. carboplatin, capecitabine)36,37,38.
We report here a gene profiling test using RNA classifiers on data from the initial biopsy to predict pCR or RD for breast cancer NAC. The BA100 model shows high negative predictive values for all subtypes of breast cancer. If BA100 was utilized to stratify patients prior to treatment, and only those predicted to have pCR were treated, pCR would then be 54% rather than 21.6% The ability to predict this response would be helpful in choosing standard options. Conversely, the ability to proactively identify a high percentage of TNBC RD patients with poor prognosis will allow physicians and patients to pursue more aggressive or targeted therapies at the beginning of treatment, rather than waiting for NAC treatment results.
Additionally, the further stratification of patients with residual disease into Class 2 and 3 with distinct biological features and significant differences in DRFS has not been detected in other recently proposed models of TNBC gene expression stratification17,27. The observation that Class 2 (lower risk RD) exhibits significantly lower expression of Ki-67 than the high-risk Class 3 or NAC-sensitive Class 1 suggests that while higher levels of proliferation may generate more patients with pCR after NAC, the tumors in Class 3 may be enriched for resistance and hence yield overall poorer outcomes. Though high levels of Ki-67 in unstratified patients were reported to respond better to NAC39, the BA100 classification system identifies high Ki-67 expressers which resist NAC and have worse outcomes than lower level Ki-67 expressers, demonstrating the value of stratifying patients. This study creates the rationale for further clinical validation and use of the test in research and development.
Data Availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Bauer, K. R., Brown, M., Cress, R. D., Parise, C. A. & Caggiano, V. Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: a population-based study from the California cancer Registry. Cancer 109, 1721–1728, https://doi.org/10.1002/cncr.22618 (2007).
Dent, R. et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res 13, 4429–4434, https://doi.org/10.1158/1078-0432.CCR-06-3045 (2007).
Kohler, B. A. et al. Annual Report to the Nation on the Status of Cancer, 1975-2011, Featuring Incidence of Breast Cancer Subtypes by Race/Ethnicity, Poverty, and State. J Natl Cancer Inst 107, djv048, https://doi.org/10.1093/jnci/djv048 (2015).
Cortazar, P. et al. Pathological complete response and long-term clinical benefit in breast cancer: the CTNeoBC pooled analysis. Lancet 384, 164–172, https://doi.org/10.1016/S0140-6736(13)62422-8 (2014).
Esserman, L. J. et al. Chemotherapy response and recurrence-free survival in neoadjuvant breast cancer depends on biomarker profiles: results from the I-SPY 1 TRIAL (CALGB 150007/150012; ACRIN 6657). Breast Cancer Res Treat 132, 1049–1062, https://doi.org/10.1007/s10549-011-1895-2 (2012).
Esserman, L. J. et al. Pathologic complete response predicts recurrence-free survival more effectively by cancer subset: results from the I-SPY 1 TRIAL–CALGB 150007/150012, ACRIN 6657. J Clin Oncol 30, 3242–3249, https://doi.org/10.1200/JCO.2011.39.2779 (2012).
Fisher, B. et al. Effect of preoperative chemotherapy on local-regional disease in women with operable breast cancer: findings from National Surgical Adjuvant Breast and Bowel Project B-18. J Clin Oncol 15, 2483–2493, https://doi.org/10.1200/JCO.1997.15.7.2483 (1997).
Fisher, B. et al. Effect of preoperative chemotherapy on the outcome of women with operable breast cancer. J Clin Oncol 16, 2672–2685, https://doi.org/10.1200/JCO.1998.16.8.2672 (1998).
Fisher, E. R. et al. Pathobiology of preoperative chemotherapy: findings from the National Surgical Adjuvant Breast and Bowel (NSABP) protocol B-18. Cancer 95, 681–695, https://doi.org/10.1002/cncr.10741 (2002).
Masood, S. Neoadjuvant chemotherapy in breast cancers. Womens Health (Lond) 12, 480–491, https://doi.org/10.1177/1745505716677139 (2016).
Papadimitriou, K., Ardavanis, A. & Kountourakis, P. Neoadjuvant therapy for locally advanced breast cancer: Focus on chemotherapy and biological targeted treatments’ armamentarium. J Thorac Dis 2, 160–170, https://doi.org/10.3978/j.issn.2072-1439.2010.02.03.8 (2010).
Ayers, M. et al. Gene expression profiles predict complete pathologic response to neoadjuvant paclitaxel and fluorouracil, doxorubicin, and cyclophosphamide chemotherapy in breast cancer. J Clin Oncol 22, 2284–2293, https://doi.org/10.1200/JCO.2004.05.166 (2004).
Hatzis, C. et al. A genomic predictor of response and survival following taxane-anthracycline chemotherapy for invasive breast cancer. JAMA 305, 1873–1881, https://doi.org/10.1001/jama.2011.593 (2011).
Lehmann, B. D. et al. Refinement of Triple-Negative Breast Cancer Molecular Subtypes: Implications for Neoadjuvant Chemotherapy Selection. PLoS One 11, e0157368, https://doi.org/10.1371/journal.pone.0157368 (2016).
Louie, M. C. & Sevigny, M. B. Steroid hormone receptors as prognostic markers in breast cancer. Am J Cancer Res 7, 1617–1636 (2017).
Mark, K. M. K., Varn, F. S., Ung, M. H., Qian, F. & Cheng, C. The E2F4 prognostic signature predicts pathological response to neoadjuvant chemotherapy in breast cancer patients. BMC Cancer 17, 306, https://doi.org/10.1186/s12885-017-3297-2 (2017).
Masuda, H. et al. Differential response to neoadjuvant chemotherapy among 7 triple-negative breast cancer molecular subtypes. Clin Cancer Res 19, 5533–5540, https://doi.org/10.1158/1078-0432.CCR-13-0799 (2013).
Nakashoji, A. et al. Clinical predictors of pathological complete response to neoadjuvant chemotherapy in triple-negative breast cancer. Oncol Lett 14, 4135–4141, https://doi.org/10.3892/ol.2017.6692 (2017).
Nwaogu, I. Y., Fayanju, O. M., Jeffe, D. B. & Margenthaler, J. A. Predictors of pathological complete response to neoadjuvant chemotherapy in stage II and III breast cancer: The impact of chemotherapeutic regimen. Mol Clin Oncol 3, 1117–1122, https://doi.org/10.3892/mco.2015.579 (2015).
Santuario-Facio, S. K. et al. A New Gene Expression Signature for Triple Negative Breast Cancer Using Frozen Fresh Tissue before Neoadjuvant Chemotherapy. Mol Med 23, https://doi.org/10.2119/molmed.2016.00257 (2017).
Tabchy, A. et al. Evaluation of a 30-gene paclitaxel, fluorouracil, doxorubicin, and cyclophosphamide chemotherapy response predictor in a multicenter randomized trial in breast cancer. Clin Cancer Res 16, 5351–5361, https://doi.org/10.1158/1078-0432.CCR-10-1265 (2010).
Denkert, C. et al. Tumour-infiltrating lymphocytes and prognosis in different subtypes of breast cancer: a pooled analysis of 3771 patients treated with neoadjuvant therapy. Lancet Oncol 19, 40–50, https://doi.org/10.1016/S1470-2045(17)30904-X (2018).
Gass, P. et al. Prediction of pathological complete response and prognosis in patients with neoadjuvant treatment for triple-negative breast cancer. BMC Cancer 18, 1051, https://doi.org/10.1186/s12885-018-4925-1 (2018).
Fournier, M. V., Fata, J. E., Martin, K. J., Yaswen, P. & Bissell, M. J. Interaction of E-cadherin and PTEN regulates morphogenesis and growth arrest in human mammary epithelial cells. Cancer Res 69, 4545–4552, https://doi.org/10.1158/0008-5472.CAN-08-1694 (2009).
Fournier, M. V. et al. Gene expression signature in organized and growth-arrested mammary acini predicts good outcome in breast cancer. Cancer Res 66, 7095–7102, https://doi.org/10.1158/0008-5472.CAN-06-0515 (2006).
Martin, K. J., Patrick, D. R., Bissell, M. J. & Fournier, M. V. Prognostic breast cancer signature identified from 3D culture model accurately predicts clinical outcome across independent datasets. PLoS One 3, e2994, https://doi.org/10.1371/journal.pone.0002994 (2008).
Lehmann, B. D. et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest 121, 2750–2767, https://doi.org/10.1172/JCI45014 (2011).
Lauterbach, N. et al. Allorecognition of HLA-DP by CD4+ T cells is affected by polymorphism in its alpha chain. Mol Immunol 59, 19–29, https://doi.org/10.1016/j.molimm.2013.12.006 (2014).
Podojil, J. R. & Miller, S. D. Potential targeting of B7-H4 for the treatment of cancer. Immunol Rev 276, 40–51, https://doi.org/10.1111/imr.12530 (2017).
Hess, K. R. et al. Pharmacogenomic predictor of sensitivity to preoperative chemotherapy with paclitaxel and fluorouracil, doxorubicin, and cyclophosphamide in breast cancer. J Clin Oncol 24, 4236–4244, https://doi.org/10.1200/JCO.2006.05.6861 (2006).
Popovici, V. et al. Effect of training-sample size and classification difficulty on the accuracy of genomic predictors. Breast Cancer Res 12, R5, https://doi.org/10.1186/bcr2468 (2010).
Miyake, T. et al. GSTP1 expression predicts poor pathological complete response to neoadjuvant chemotherapy in ER-negative breast cancer. Cancer Sci 103, 913–920, https://doi.org/10.1111/j.1349-7006.2012.02231.x (2012).
Symmans, W. F. et al. Measurement of residual breast cancer burden to predict survival after neoadjuvant chemotherapy. J Clin Oncol 25, 4414–4422, https://doi.org/10.1200/JCO.2007.10.6823 (2007).
Rouzier, R. et al. Breast cancer molecular subtypes respond differently to preoperative chemotherapy. Clin Cancer Res 11, 5678–5685, https://doi.org/10.1158/1078-0432.CCR-04-2421 (2005).
Wallden, B. et al. Development and verification of the PAM50-based Prosigna breast cancer gene signature assay. BMC Med Genomics 8, 54, https://doi.org/10.1186/s12920-015-0129-6 (2015).
Carey, L. A. et al. The triple negative paradox: primary tumor chemosensitivity of breast cancer subtypes. Clin Cancer Res 13, 2329–2334, https://doi.org/10.1158/1078-0432.CCR-06-1109 (2007).
Chan, J. J., Tan, T. J. Y. & Dent, R. A. Are There Any Clinically Relevant Subgroups of Triple-Negative Breast Cancer in 2018? J Oncol Pract 14, 281–289, https://doi.org/10.1200/JOP.18.00034 (2018).
Costa, R. L. B. & Gradishar, W. J. Triple-Negative Breast Cancer: Current Practice and Future Directions. J Oncol Pract 13, 301–303, https://doi.org/10.1200/JOP.2017.023333 (2017).
Wang, R. X., Chen, S., Jin, X. & Shao, Z. M. Value of Ki-67 expression in triple-negative breast cancer before and after neoadjuvant chemotherapy with weekly paclitaxel plus carboplatin. Sci Rep 6, 30091, https://doi.org/10.1038/srep30091 (2016).
Acknowledgements
We gratefully acknowledge the support for the I-SPY1 TRIAL provided by the National Cancer Institute Specialized Program of Research Excellence in breast cancer, American College of Radiology Imaging Network, Cancer and Leukemia Group B, National Cancer Institute Center for Bioinformatics, and the Breast Cancer Research Foundation. The research has been funded by Bioarray Genetics Inc.
Author information
Authors and Affiliations
Contributions
M.V.F., E.C.G., J.C., J.O., S.H.T., and A.M.B. contributed with conception, experimental design, analyses, and co-authoring the manuscript.
Corresponding author
Ethics declarations
Competing Interests
MVF, is an employee, inventor, and owns stock options of Bioarray Genetics Inc. ECG, is a former employee and is an investor and owns stock options of Bioarray Genetics Inc. JCO is an employee of Rancho Biosciences. JC is a former consultant for Rancho Biosciences and a current consultant for Bioarray Genetics Inc. SHT, and AMB are advisors with stock options of Bioarray Genetics Inc.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Fournier, M.V., Goodwin, E.C., Chen, J. et al. A Predictor of Pathological Complete Response to Neoadjuvant Chemotherapy Stratifies Triple Negative Breast Cancer Patients with High Risk of Recurrence. Sci Rep 9, 14863 (2019). https://doi.org/10.1038/s41598-019-51335-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-51335-1
This article is cited by
-
Basal–epithelial subpopulations underlie and predict chemotherapy resistance in triple-negative breast cancer
EMBO Molecular Medicine (2024)
-
Predicting pathological complete response after neoadjuvant chemotherapy in breast cancer by clinicopathological indicators and ultrasound parameters using a nomogram
Scientific Reports (2024)
-
Decoding gene regulatory circuitry underlying TNBC chemoresistance reveals biomarkers for therapy response and therapeutic targets
npj Precision Oncology (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.