Abstract
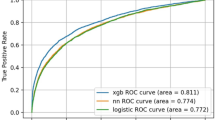
A comprehensive screening method using machine learning and many factors (biological characteristics, Helicobacter pylori infection status, endoscopic findings and blood test results), accumulated daily as data in hospitals, could improve the accuracy of screening to classify patients at high or low risk of developing gastric cancer. We used XGBoost, a classification method known for achieving numerous winning solutions in data analysis competitions, to capture nonlinear relations among many input variables and outcomes using the boosting approach to machine learning. Longitudinal and comprehensive medical check-up data were collected from 25,942 participants who underwent multiple endoscopies from 2006 to 2017 at a single facility in Japan. The participants were classified into a case group (y = 1) or a control group (y = 0) if gastric cancer was or was not detected, respectively, during a 122-month period. Among 1,431 total participants (89 cases and 1,342 controls), 1,144 (80%) were randomly selected for use in training 10 classification models; the remaining 287 (20%) were used to evaluate the models. The results showed that XGBoost outperformed logistic regression and showed the highest area under the curve value (0.899). Accumulating more data in the facility and performing further analyses including other input variables may help expand the clinical utility.
Similar content being viewed by others
Introduction
Gastric cancer is one of the most common cancers in the world, and Japan has one of the highest incidences1,2. Gastric cancer also has high mortality rates in East Asian countries3,4. Long-term Helicobacter pylori infection, pernicious anaemia and high salt intake can lead to chronic superficial gastritis, chronic atrophic gastritis and eventually intestinal epithelial metaplasia, all of which are considered risk factors for the development of gastric cancer5,6,7.
It is important to provide accurate, rapid screening for gastric cancer. If a patient is predicted as being at high risk, then (s)he can seek to undertake preventative measures in advance. Conversely, if a patient is predicted as being at low risk, then (s)he can avoid or reduce the frequency of (e.g. annually in Japan) upper gastrointestinal endoscopic examinations, which are accompanied by potential risks and high screening costs. A large-scale survey of 200,000 individuals who had been endoscopically examined reported a 0.13% adverse complication rate and a 0.004% mortality rate8. Therefore, endoscopic gastric cancer screening has been proposed in several subgroups of patients considered to be at high risk9.
While various environmental risk and host-related factors have been suggested to be associated with gastric cancer, rapid screening to classify patients as high or low risk of developing gastric cancer in the clinical setting is often provided based on a few main factors: age, familial history and the presence of H. pylori infection or atrophic gastritis.
Some recent studies have demonstrated that new methods such as machine learning and big data mining approaches are effective for improving screening, prediction, biomarker selection and disease diagnosis in the medical field10,11,12,13,14,15. We hypothesized that comprehensive screening using a combination of numerous factors accumulated every day in hospitals (e.g. biological characteristics, H. pylori infection status, endoscopic findings and blood test results) and a successful machine learning technique could lead to more accurate and rapid screening for gastric cancer.
One such advanced and successful machine learning method is XGBoost16,17. XGBoost uses multiple (hundreds of) classification and regression trees (CARTs), which can learn nonlinear relations among input variables and outcomes in a boosting ensemble manner, to capture and learn nonlinear and complex relations accurately (see the XGBoost subsection for technical details). Linear approaches such as logistic regression are not generally suitable for prediction models with complex correlations; however, multiple risk factors may jointly and nonlinearly help predict the risk of developing gastric cancer. Therefore, the purpose of the present study was to clarify the accuracy of a prediction model for the development of gastric cancer using comprehensive longitudinal data and machine learning algorithms.
Results
We considered a classification problem regarding whether a subject would have a future risk of gastric cancer by predicting whether (s)he would be diagnosed with gastric cancer within the next 122 months. To study this, we collected longitudinal and comprehensive medical check-up data from 25,942 participants who underwent multiple endoscopies from 2006 to 2017 at a single facility in Japan (see the Methods section for details of the data collection). We classified the participants into a case group (y = 1) or a control group (y = 0) if gastric cancer was or was not detected, respectively, during the 122-month period. As a result, 1,431 participants (89 cases and 1,342 controls) were extracted. From the participants, 1,144 (80%) were randomly selected for use in training classification models, and the remaining 287 (20%) were used to evaluate the prediction accuracy of the constructed models. Classification performance was measured by receiver operating characteristic (ROC) curves and their area under the curve (AUC) values. In addition to the ROC and AUC values, the resulting accuracy, sensitivity, specificity and its confusion matrix determined by a cut-off value of 0.5 were reported. We constructed 10 classification models to address the following two research questions. Table 1 shows a list of the 10 constructed classification models (models A–J) using XGBoost and logistic regression, while incrementally adding input variables related to risk factors of gastric cancer (see the Statistical analysis subsection for details of the input variables).
Increasing input variables to predict future gastric cancer
Questions
Long-term H. pylori infection and the presence of chronic atrophic gastritis are known risk factors for the future development of gastric cancer, and clinicians also often take the patient’s state into consideration. Our first question was whether only these two factors are sufficient to predict future gastric cancer. We aimed to compute the advantages that could be obtained by adding other medical check-up information in light of an advanced machine learning technique.
Results
Figure 1 shows the resulting five ROC curves for models A–E obtained using the XGBoost technique. The horizontal and vertical axes represent the false and true positive rates, respectively. Model A, which inputs solely the presence of H. pylori infection, is indicated by the light blue line. Model B, which inputs the presence of both H. pylori infection and chronic atrophic gastritis, is indicated by the green line. Model C, which added other endoscopic findings to model B, is indicated by the pink line. Model D, which added biological background factors to model C, is indicated by the red line. Finally, model E, which added blood test results to model D, is indicated by the black line. We see that the classification performance was increased by adding input variables. Especially, models D and E showed a significant increase by adding age, body mass index (BMI), and blood test results. Table 2 presents the AUC values for the cross-validation (CV) and test data corresponding to Fig. 1, along with the accuracy, sensitivity and specificity. AUC values generally increased with increasing numbers of input variables. Model E, which exploited all the information for the input variables, showed the best AUC value (0.899) for the unknown test data, with accuracy = 0.777, sensitivity = 0.933 and specificity = 0.768. In addition, the confusion matrix (true positive, false negative, false positive, true negative) for each model is shown in Supplementary Table S1. Models D and E predicted a small number of patients as false negative, which was a favourable result for a rapid screening method.
In general, machine learning methods possess hyperparameters that should be carefully selected by practitioners. Different hyperparameter settings may yield different classification results. XGBoost possesses several hyperparameters (see the Hyperparameter tuning section for details) that should be optimized using training data. To optimize the hyperparameters, we took advantage of the Bayesian optimization (BO) method18,19, which enables the automatic selection of a favourable combination of hyperparameters using Gaussian process regression. Supplementary Figs S1 to S5 show the time course of AUC values on the CV (XGB_cv) and test data (XGB_test) as a function of XGBoost hyperparameter updates by BO. Whereas the AUC value (XGB_cv) was monotonically increasing, the AUC value (XGB_test) was not. One of the reasons for this could be the limited amount of test data. However, models A–E were observed to have a robust ranking order in terms of AUC values under several hyperparameter updates.
Lastly, Supplementary Table S2 presents importance scores for each input variable provided by XGBoost, which contribute to the prediction of future gastric cancer. In model E, HbA1c, mean corpuscular volume (MCV), lymphocyte ratio, age, BMI and post-gastrectomy were found to be more important variables. Sex was not an important variable. The automatically computed risk factors for gastric cancer were reasonable, as explained in the Discussion section below.
Findings
Two known risk factors, long-term H. pylori infection and the presence of chronic atrophic gastritis, could not accurately predict future gastric cancer. Adding other factors, biological background and blood test results, led to increased classification performance in terms of the ROC and AUC values. Important input variables automatically selected by XGBoost agreed with previous reports, as described in the Discussion section.
Nonlinear machine learning versus linear logistic regression
Questions
Our second question was whether an advanced and successful nonlinear machine learning method (XGBoost) could be effective for predicting the future gastric cancer compared with a traditional linear method (logistic regression). We aimed to compute the advantages that could be obtained using XGBoost compared with linear logistic regression.
Results
We compared the nonlinear machine learning technique (XGBoost) with the linear method (logistic regression) as a baseline. In Table 2, models F–J show the results of linear logistic regression, which used the same input variables as XGBoost (Table 1). We found that XGBoost outperformed logistic regression when the input variables were increased (models D and I, models E and J). Whereas models A, B, C, F, G and H input only binary variables, models D, E, I and J input many continuous variables. XGBoost was able to capture the nonlinear relations between the outcome and many input variables by learning, and showed superior performance compared with traditional logistic regression. Supplementary Figs S1 to S5 show a comparison between XGBoost and logistic regression in terms of the time course of AUC values under hyperparameter updates by BO. Although XGBoost and logistic regression showed almost the same results in a few input variables (models A, B, C, F, G and H), they showed significant differences when increasing the number of input variables (models D, E, I and J).
Findings
In the task of predicting future gastric cancer risk, the relation between the outcome and many existing clinical markers (Table 1) was nonlinear; thus, by capturing the nonlinearity, XGBoost achieved higher performance than linear logistic regression. Increasing the number of input factors related to gastric cancer and utilizing advanced nonlinear machine learning methods appears to be effective to predict future gastric cancer risk accurately.
Discussion
Four gastric cancer screening methods are generally performed: upper gastrointestinal series, H. pylori serology, serum pepsinogen test and endoscopy. It has been reported that a combination of anti-H. pylori antibody and serum pepsinogen can accurately predict the development of gastric cancer20, and in Japan, gastric cancer screening is actually performed using this combination. In a previous review, Kim et al.21 suggested that gastric cancer screening in the U.S. should be stratified by region, age, family history of gastric cancer, H. pylori infection and gastric conditions (atrophic gastritis/intestinal metaplasia). Comprehensive assessments of multiple risk factors may contribute to the improved accuracy of gastric cancer screening, and previous reports have suggested other factors that may be associated with the risk of developing gastric cancer, such as diabetes and obesity22,23. Pernicious anaemia has also been suggested to be associated with the presence of gastric cancer24. In addition, differential leukocyte count, including lymphocyte count, may be useful in predicting prognosis or the presence of gastric cancer25,26. The risk of gastric cancer may increase after gastric surgery27. In our comprehensive prediction models, especially model E, the indicators related to these previous reports—HbA1c, MCV, lymphocyte ratio, age, BMI and post-gastrectomy—contributed to our calculations; this resulted in higher accuracy compared with prediction model B, which used merely information regarding H. pylori infection and the presence of atrophic gastritis. This suggests that calculations using comprehensive data with an advanced nonlinear machine learning method improve prediction accuracy.
Our results may provide information for classifying “high-risk patients” who should be recommended for frequent endoscopic screening for gastric cancer, and “low-risk patients” who should not. Few studies have been conducted on the optimal intervals for endoscopic gastric cancer screening, and no guidelines currently exist. Gastric cancer screening is recommended every 1–2 years for high-risk patients in many countries21. On the other hand, in their review, Kim et al.21 recommend endoscopic gastric cancer screening every 3–5 years for low-risk patients in the U.S. With reference to these previous reports, if an accurate screening test is obtained after expanding and improving the present study, we may recommend gastric cancer screening every 1–2 years for high-risk patients and every 3–5 years for low-risk patients.
We defined patients for whom no gastric cancer could be detected for 122 months or longer as the control group. We set 122 months as a cut-off because it is the longest period that gastric cancer could be detected in the case group. It is generally thought that cancer initiation begins about 20 years before detection. Thus, the control group may have included patients who have the potential to develop gastric cancer in the future. The control group in our study did not comprise “patients who will not develop gastric cancer”, but rather “patients in whom gastric cancer will not be detected within a few years”.
Our research had several limitations. First, in our survey, we did not use information obtained from questionnaires regarding the participants’ dietary habits. It is widely known that long-term consumption of a high-salt diet is a risk factor for gastric cancer; however, our questionnaire did not collect information on salt consumption. In addition, various other environmental risk factors (e.g. smoking) and host-related factors (e.g. blood type) have been suggested to be associated with gastric cancer. Although such data were not available this time, it seems that with more related variables included, the model performance would increase.
Second, incidentally, some patients without H. pylori infection or atrophic gastritis were classified into the case group; the possible reasons for this are described below. It is widely known that patients in the end stage of chronic atrophic gastritis can show conversion from H. pylori-positive to -negative. The case group also included patients who received therapy for H. pylori eradication. Because our study used the initial examination data, patients with early gastric findings that were not identified may have been included in the case group.
Third, it remains unclear whether the results from our prediction can be generalized to other populations. Our predictions were calculated based on data from 1,431 patients (89 cases and 1,342 controls). It takes approximately 10 years to obtain one (control) sample for the long-term (122 months) prediction of the development of gastric cancer. We are concerned that the amount of data collected is insufficient to generalize our results. Therefore, collaborative multicentre research would be helpful.
Finally, in our study, almost all of the endoscopic findings, including part of the gastric cancer diagnoses, were diagnosed based on macroscopic findings by endoscopists; therefore, these endoscopic findings may include errors and biases. Mechanical diagnoses of stomach findings (such as the deep learning method28 using upper gastrointestinal endoscopic images) may be able to help avoid human errors and biases. Combining deep-learning-based diagnoses and comprehensive screening methods as in the present study is planned for our future research.
Methods
Study design and population
This study was conducted as a retrospective, observational, single-centre study. Most of the participants were aged between 40–60 years, and were primarily healthy clerical staff and volunteers from among the employees of the Nippon Telegraph and Telephone Corporation (NTT) and their family members. The study data were obtained from a comprehensive, periodic health check-up program carried out at the Center for Preventive Medicine, NTT Medical Center Tokyo, from May 2006 to November 2017. In Japan, the Occupational Health and Safety Law requires employers to provide annual health check-ups to ensure the health of their employees. To comply with this law, the Center for Preventive Medicine has been contracted by NTT to provide periodic medical examinations to their employees. This program includes a comprehensive periodic medical examination and many more services than required by law; the data used in this study were collected at the Center for Preventive Medicine as part of this general health check-up program. We did not specifically intend to collect new data for this study, so we extracted the participants’ past clinical data from the institution’s database. The research plan was described on the websites of both our facility and the Center for Preventive Medicine. It was announced that participants could withdraw from the study at any time without negative consequences. As this was a retrospective study, the need for informed consent was waived by the review board. The study protocol was approved by the ethics review board of Juntendo University (No. 2018148) and the institutional ethics committee at the Center for Preventive Medicine (No. 18-106). All procedures were performed in accordance with the relevant guidelines and regulations.
Data collection
Each participant’s weight and height were measured after they removed their shoes and heavy clothing. BMI was calculated as weight in kilograms divided by height in metres squared (kg/m2). Serum samples were collected from each participant after overnight fasting and immediately subjected to biochemical analysis. The Japan Diabetes Society (JDS) HbA1c values were converted to National Glycohemoglobin Standardization Program values using the formula developed by the JDS: HbA1c = [HbA1c(JDS)(%) × 1.02 + 0.25(%)]29. Screening using esophagogastroduodenoscopy was performed by endoscopy specialists belonging to the NTT Medical Center Tokyo. A diagnosis of gastric cancer was given when gastric cancer was strongly suspected by the endoscopic findings, or when adenocarcinoma was identified in biopsy specimens. Diagnosis of H. pylori infection was based on the detection of H. pylori-specific antibodies in serum. H. pylori-specific immunoglobulin G antibody titres were measured using a commercially available enzyme-linked immunosorbent assay (ELISA) according to the manufacturer’s instructions (Eiken Chem Corp., Tokyo, Japan). Participants with antibody titres 10 U/mL or higher were considered seropositive for H. pylori infection30. The sensitivity and specificity of the assay for H. pylori infection have been reported as follows: sensitivity 88% and <95% and specificity 84% and <95%, respectively31,32. Our investigation focused on 25,942 patients who underwent upper gastrointestinal endoscopy more than once between May 2006 and November 2017. Supplementary Table S3 shows the distribution of patients by the number of esophagogastroduodenoscopies obtained during the examination period. Overall, 85 patients underwent esophagogastroduodenoscopies twice, and one patient 14 times. During that period, 89 patients were diagnosed with gastric cancer, with 76 being identified as having adenocarcinoma based on biopsy specimens, and 13 strongly suspected of having gastric cancer based on endoscopic findings. The maximum examination period until the development of gastric cancer identified in the 89 patients was 122 months.
Statistical analysis
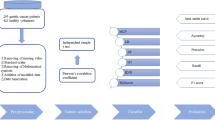
Data from each participant’s first health check-up were used to predict whether (s)he would later be diagnosed with gastric cancer during the examination period. To apply a supervised machine learning method such as XGBoost, a data label (case: y = 1, control: y = 0) is required for each participant. We defined the case group (y = 1) as patients with detected gastric cancer within 122 months (89 patients), and the control group (y = 0) as the negation: patients with no detected gastric cancer at 122 months or longer (1,342 patients). The participants could be classified into three subject categories: X, Y and Z. Subject X was for those in whom gastric cancer was detected in the second or subsequent endoscopic examinations (case group). Subject Y was for those in whom gastric cancer was not detected by endoscopy multiple times at more than or equal to 122 months (control group). Subject Z was for those in whom no gastric cancer was detected; those examined at intervals less than 122 months were excluded. Figure 2 illustrates this classification.
Table 3 shows the summary statistics of the analysed initial medical check-up data of the case and control groups. The patients in the case group were older and showed a higher prevalence of H. pylori infection, upper gastrointestinal ulcer and chronic atrophic gastritis. In the serologic examination, the case group had higher MCV and HbA1c values. Figure 3 shows the inclusion and exclusion procedures in our analysis. Overall, 24,511 patients with a follow-up period of less than 122 months were excluded. Finally, the initial examination data from 1,431 patients were analysed. We trained classification models by using randomly selected training data (1,144 patients [80%]), and evaluated the prediction accuracy on the remaining test data (287 patients [20%]). To predict the future risk of gastric cancer, we developed 10 classification models with different input variables (A–J), as shown in Table 1. Model A inputs information regarding H. pylori infection as a leading cause of gastric cancer4,33. Model B additionally inputs the presence of atrophic gastritis, one of the main risk factors for gastric cancer34,35. Model C additionally inputs other endoscopic findings considered to be risk factors for gastric cancer, including gastric ulcer (scar), duodenal ulcer (scar) and gastrectomy36,37,38. Model D additionally inputs biological characteristics such as sex, age and BMI, as obesity has been reported to be a risk factor for gastric cancer22. Model E additionally inputs blood test variables (complete blood count and HbA1c), as pernicious anaemia may be associated with an increased risk of gastric cancer24,39. In addition, the possibility that diabetes is associated with gastric cancer has been reported23,40. Compared with XGBoost, models F–J were calculated using the same variables by logistic regression.
XGBoost
XGBoost open source software16,17 provides machine learning solutions to classification and regression tasks using techniques of ensemble learning with gradient tree boosting (GTB)41. XGBoost is well known for obtaining winning solutions in various data competitions. Chen and Guestrin16 reported that “among the 29 challenge-winning solutions published on Kaggle’s blog during 2015, 17 winning solutions used XGBoost.” Practical applications of XGBoost include “store sales prediction, high energy physics event classification, Web text classification, customer behaviour prediction, motion detection, ad click-through rate prediction, malware classification, product categorization, hazard risk prediction and massive on-line course dropout rate prediction.” XGBoost has also been applied to the medical field10,11,12. We used XGBoost to classify subjects as high or low risk in terms of developing gastric cancer. XGBoost (or GTB) can capture nonlinear relations among the outcome and input variables by sequentially learning multiple CARTs; this jointly helps detect early signs of gastric cancer. CARTs possess parameters that need to be learned, such as various tree structure options and leaf node weights. XGBoost automatically learns multiple CARTs with these parameters by optimising a loss function using gradient methods. In addition, XGBoost includes several hyperparameters that need to be tuned, as described below.
Evaluation
A trained classification model outputs a probability value [0–1] of the risk of developing gastric cancer for each participant. To evaluate the 10 trained classification models, we used ROC curves and their AUC values as computed from the test data42,43. ROC curves have commonly been used to measure classification performance. We report a confusion matrix (true positive, false negative, false positive, true negative) at the cut-off value determined by the probability value of 0.5 (i.e. if the probability value ≥0.5, then the patient is classified as being at risk; otherwise as low-risk).
Hyperparameter tuning: bayesian optimization (BO)
XGBoost includes several hyperparameters that need to be tuned, including the maximum depth of regression trees, number of weak learners (CARTs), subsample ratios of columns and training instances for constructing each tree, partitioning-leaf-node parameters (gamma), imbalance parameters, learning rate (eta) and regularization parameters (lambda and alpha). The hyperparameter settings affect the performance of XGBoost. We optimized the hyperparameters to maximize the mean AUC value computed from 5-fold CV on the training data. Specifically, the training data were randomly divided into five subsets: four were used for training XGBoost and the other was used for validation. An ROC curve and AUC value could be evaluated from the validation subset. This procedure was repeated five times with different validation subsets, and then the mean AUC value was computed by averaging the five AUC values. We used the BO method18,19 to maximize automatically the mean AUC value (AUC-CV). BO iteratively suggests favourable hyperparameter values that effectively increase the objective function (AUC-CV) by learning the previously observed pairs of hyperparameters and their AUC-CV values. After identifying the favourable hyperparameter values, XGBoost was trained using the entire training dataset. The final ROC and AUC values were then evaluated using the test data (AUC-Test). Supplementary Figs S1 to S5 show the time courses of the monotonically increased AUC-CV values and resulting AUC-Test values for each model A–J as a function of iterations of BO explorations for hyperparameters. XGBoost and logistic regression showed a significant difference in models D, E, I and J with many input variables.
Data Availability
The datasets generated during and/or analysed during the current study are not publicly available because of access to information being severely restricted by the ethics committee of NTT Medical Center Tokyo, but may be available from the corresponding author on reasonable request.
References
Jemal, A. et al. Global cancer statistics. CA Cancer J. Clin. 61, 69–90, https://doi.org/10.3322/caac.20107. Erratum in: CA Cancer J. Clin. 61, 134 PMID: 21296855 (2011).
GLOBOCAN 2012: Estimated cancer incidence, mortality and prevalence worldwide in 2012. World Health Organization, Geneva, Switzerland. Avaoilable online at, http://globocan.iarc.fr/Pages/Map.aspx.
Nashimoto, A. et al. Gastric cancer treated in 2002 in Japan: 2009 annual report of the JGCA nationwide registry. Gastric Cancer 16, 1–27, https://doi.org/10.1007/s10120-012-0163-4. PubMed PMID: 22729699; PMCID: PMC3549249 (2013).
Ferlay, J. et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 136, E359–386, https://doi.org/10.1002/ijc.29210. PMID: 25220842 (2015).
Correa, P. A human model of gastric carcinogenesis. Cancer Res. 48, 3554–3560 PMID: 3288329 (1988).
Schistosomes, liver flukes and Helicobacter pylori. IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Lyon, 7–14 June 1994. IARC Monogr. Eval. Carcinog. Risks Hum. 61, 1–241 PMID: 7715068 (1994).
Tsugane, S. & Sasazuki, S. Diet and the risk of gastric cancer: review of epidemiological evidence. Gastric Cancer 10, 75–83 PMID: 17577615 (2007).
Silvis, S. E., Nebel, O., Rogers, G., Sugawa, C. & Mandelstam, P. Endoscopic complications. Results of the 1974 American Society for Gastrointestinal Endoscopy Survey. JAMA 235, 928–930 PMID: 128642 (1976).
ASGE Standards of Practice Committee et al. The role of endoscopy in the management of premalignant and malignant conditions of the stomach. Gastrointest. Endosc. 82, 1–8, https://doi.org/10.1016/j.gie.2015.03.1967. PMID: 25935705 (2015).
Gao, C. et al. Model-based and model-free machine learning techniques for diagnostic prediction and classification of clinical outcomes in Parkinson’s disease. Sci Rep. 8, 7129 PMID: 29740058 (2018).
Nishio, M. et al. Computer-aided diagnosis of lung nodule using gradient tree boosting and Bayesian optimization. PLoS One 13, e0195875 PMID: 29672639 (2018).
Qiao, Z. et al. Using machine learning approaches for emergency room visit prediction based on electronic health record data. Stud. Health Technol. Inform. 247, 111–115 PMID: 29677933 (2018).
Hinton, D. J. et al. Metabolomics biomarkers to predict acamprosate treatment response in alcohol-dependent subjects. Sci Rep. 7, 2496 PMID: 28566752 (2017).
Liu, R. Zhang, G., & Yang, Z. Towards rapid prediction of drug-resistant cancer cell phenotypes: single cell mass spectrometry combined with machine learning. Chem Commun. (Camb) 55, 616–619 PMID: 30525135 (2019).
Maeta, K. et al. Prediction of glucose metabolism disorder risk using a machine learning algorithm: pilot study. JMIR Diabetes 26, e10212 PMID: 30478026 (2018).
Chen, T. & Guestrin, C. XGBoost: a scalable tree boosting system. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (KDD’ 16). ACM, New York, NY, USA, 785–794 (2016).
GitHub: XGBoost, Distributed (Deep) Machine Learning Community. Available online at, https://github.com/dmlc/xgboost.
Brochu, E., Cora, V. M. & de Freitas, N. A tutorial on Bayesian optimization of expensive cost functions, with application to active user modeling and hierarchical reinforcement learning. CoRR. arXiv 1012, 2599 (2010).
Shahriari, B., Swersky, K., Wang, Z., Adams, R. P. & de Freitas, N. Taking the human out of the loop: a review of Bayesian optimization. Proc. IEEE 104, 148–175 (2016).
Watabe, H. et al. Predicting the development of gastric cancer from combining Helicobacter pylori antibodies and serum pepsinogen status: a prospective endoscopic cohort study. Gut 54, 764–768 PubMed PMID: 15888780; PMCID: PMC1774550 (2005).
Kim, G. H., Liang, P. S., Bang, S. J. & Hwang, J. H. Screening and surveillance for gastric cancer in the United States: Is it needed? Gastrointest. Endosc. 84, 18–28 PMID: 26940296 (2016).
Yang, P. et al. Overweight, obesity and gastric cancer risk: results from a meta-analysis of cohort studies. Eur. J. Cancer 45, 2867–2873 PMID: 19427197 (2009).
Inoue, M. et al. Diabetes mellitus and the risk of cancer: results from a large-scale population-based cohort study in Japan. Arch. Intern. Med. 166, 1871–1877 PubMed PMID: 17000944 (2006).
Vannella, L., Lahner, E., Osborn, J. & Annibale, B. Systematic review: gastric cancer incidence in pernicious anaemia. Aliment. Pharmacol. Ther. 37, 375–382 PMID: 23216458 (2013).
Feng, F. et al. Low lymphocyte count and high monocyte count predicts poor prognosis of gastric cancer. BMC Gastroenterol. 18, 148 PMID: 30305076; PMCID: PMC6180580 (2018).
Jiang, Y., Xu, H., Jiang, H., Ding, S. & Zheng, T. Pretreatment neutrophil-lymphocyte count ratio may associate with gastric cancer presence. Cancer Biomark. 16, 523–528 PMID: 27062567 (2016).
Takeno, S. et al. Gastric cancer arising from the remnant stomach after distal gastrectomy: a review. World J. Gastroenterol. 20, 13734–13740 PMID: 25320511; PMCID: PMC4194557 (2014).
Itoh, T., Kawahira, H., Nakashima, H. & Yata, N. Deep learning analyzes Helicobacter pylori infection by upper gastrointestinal endoscopy images. Endosc. Int. Open 6, E139–E144 PMID: 29399610; PMCID: PMC5794437 (2018).
Committee on the Standardization of Diabetes Mellitus-Related Laboratory Testing of Japan Diabetes Society. International clinical harmonization of glycated hemoglobin in Japan: From Japan Diabetes Society to National Glycohemoglobin Standardization Program values. J. Diabetes Investig. 3, 39–40 PMID: 24843544 (2012).
Kikuchi, S. & Miwa, H. Evaluation of direct ELISA kit (E-plate) in diagnosing Helicobacter pylori infection. Med. Pharmacol. (in Japanese) 43, 581–586 (2000).
Longo-Mbenza, B., Nkondi Nsenga, J. & Vangu Ngoma, D. Prevention of the metabolic syndrome insulin resistance and the atherosclerotic diseases in Africans infected by Helicobacter pylori infection and treated by antibiotics. Int. J. Cardiol. 121, 229–238 PMID: 17368586 (2007).
Woodward, M., Morrison, C. & McColl, K. An investigation into factors associated with Helicobacter pylori infection. J. Clin. Epidemiol. 53, 175–181 PMID: 10729690 (2000).
Correa, P. et al. Helicobacter pylori and gastric carcinoma. Serum antibody prevalence in populations with contrasting cancer risks. Cancer 66, 2569–2574 PMID: 2249197 (1990).
Genta, R. M. Acid suppression and gastric atrophy: sifting fact from fiction. Gut 43, Suppl 1: S35–S38 PMID: 9764038; PMCID: PMC1766607 (1998).
Tatsuta, M. et al. Fundal atrophic gastritis as a risk factor for gastric cancer. Int. J. Cancer 53, 70–74 PMID: 8416206 (1993).
Takeno, S. et al. Gastric cancer arising from the remnant stomach after distal gastrectomy: a review. World J. Gastroenterol. 20, 13734–13740 PMID: 25320511; PMCID: PMC4194557 (2014).
Neugut, A. I., Hayek, M. & Howe, G. Epidemiology of gastric cancer. Semin. Oncol. 23, 281–291 PMID: 8658212 (1996).
Hansson, L. E. et al. The risk of stomach cancer in patients with gastric or duodenal ulcer disease. N. Engl. J. Med. 335, 242–249 PMID: 8657240 (1996).
Landgren, A. M. et al. Autoimmune disease and subsequent risk of developing alimentary tract cancers among 4.5 million US male veterans. Cancer 117, 1163–1171 PMID: 21381009; PMCID: PMC3052786 (2011).
Hemminki, K., Li, X., Sundquist, J. & Sundquist, K. Risk of cancer following hospitalization for type 2 diabetes. Oncologist 15, 548–555 PubMed PMID: 20479278; PMCID: PMC3227990 (2010).
Friedman, J. H. Greedy function approximation: a gradient boosting machine. Ann. Stat. 29, 1189–1232 (2001).
Metz, C. E. Receiver operating characteristic analysis: a tool for the quantitative evaluation of observer performance and imaging systems. J. Am. Coll. Radiol. 3, 413–422 PMID: 17412096 (2006).
Krzanowski, W. J. & Hand, D. J. ROC curves for continuous data, monographs on statistics and applied probability 111. (Chapman and Hall/CRC Press, Boca Raton, FL 2009).
Acknowledgements
We thank Forte Science Communications, Inc. for editorial assistance with a draft of this manuscript. This study was funded in part by a grant received by Kazutoshi Fujibayashi from Novartis Pharma K.K. (no reference number).
Author information
Authors and Affiliations
Contributions
The experiments were conceived and designed by F.K., N.Y., G.T. and N.T.; the experiments were carried out by T.J., N.Y., F.K. and G.T.; S.N., I.K., G.T., F.K., T.J. and N.Y. contributed significantly to the execution, and/or analysis and interpretation of the data; the paper was written by F.K., N.Y. and T.J.; I.K., S.N., G.T. and N.T. participated in reviewing and/or revising the manuscript for intellectual content; F.K., T.J., N.Y., I.K., S.N., G.T. and N.T. approved the manuscript for submission.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Taninaga, J., Nishiyama, Y., Fujibayashi, K. et al. Prediction of future gastric cancer risk using a machine learning algorithm and comprehensive medical check-up data: A case-control study. Sci Rep 9, 12384 (2019). https://doi.org/10.1038/s41598-019-48769-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-48769-y
This article is cited by
-
Risk prediction model for gastric cancer within 5 years in healthy Korean adults
Gastric Cancer (2024)
-
Interpretable machine learning model integrating clinical and elastosonographic features to detect renal fibrosis in Asian patients with chronic kidney disease
Journal of Nephrology (2024)
-
Early prediction of response to palliative chemotherapy in patients with stage-IV gastric and esophageal cancer
BMC Cancer (2023)
-
Establishing machine learning models to predict the early risk of gastric cancer based on lifestyle factors
BMC Gastroenterology (2023)
-
An ensemble method of the machine learning to prognosticate the gastric cancer
Annals of Operations Research (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.