Abstract
Topoisomerases II (Top2s) are a group of essential enzymes involved in replication, transcription, chromosome condensation, and segregation via altering DNA topology. The mechanism of the Top2s poisons such as etoposide (VP-16) was reported as stabilizing the Top2-DNA complex and engendering permanent DNA breakage. As the structurally similar compound of VP-16, a novel 4β-sulfur-substituted 4′-demethylepipodophyllotoxin (DMEP) derivative (compound C-Bi) with superior antitumor activity was developed in our previous study. To understand the structural basis of the compound action, the crystal structure (2.54 Å) of human Top2 β-isoform (hTop2β) cleavage complexes stabilized by compound C-Bi was determined. However, compound C-Bi was not visible in the crystal structure. Through the comparison of the structures of hTop2β-DNA-etoposide ternary complex and hTop2β-DNA binary complex, it could be observed that the distance between drug-binding sites Arg503 of the two monomers was 26.62 Å in hTop2β-DNA-etoposide ternary complex and 34.54 Å in hTop2β-DNA binary complex, respectively. Significant twist were observed in the DNA chains of binary complex. It suggested that compound C-Bi played antitumor roles through increasing spacing of hTop2β monomers. The changes in hTop2β structure further caused double changes in the torsional direction and migration distance of the DNA chains, resulting in impeding religation of DNA.
Similar content being viewed by others
Introduction
Topoisomerases II (Top2s) are a group of essential nuclear enzymes that alter the topological structure of DNA through the transient cleavage, strand passage and subsequent re-ligation of double-stranded DNA1,2,3,4. Top2s in mammalian cells can be divided into two subtypes which are type IIα (Top2α) and IIβ (Top2β)5. Top2α plays roles in proliferating cells and assists in replication, chromosome condensation and segregation6,7. And Top2β is associated with DNA repair and transcription2,8. Especially, site-specific cleavage of DNA by human topoisomerases IIβ (hTop2β) has been reported to be necessary to activate the transcription of some genes9.
Beyond the vital cellular functions, Top2s are considered as an important target protein for some of the most active drugs for the treatment of human cancers10,11,12. Top2s-targeting drugs, such as etoposide (VP-16), are among the most effective and widely used anticancer drugs in cancer chemotherapy13,14,15. These drugs are referred to as Top2s poisons, because they convert Top2s into a physiological toxin that creates DNA double-strand breaks by increasing the steady-state levels of DNA cleavage complex16,17. Top2s-mediated DNA damage can further activate a series of intracellular signal response, and ultimately lead to tumor cells death18,19. To understand the structural basis of Top2s-targeting drug, especially Top2β-targeting, the high-resolution crystal structure of a ternary VP-16-hTop2β catalytic core-DNA complex has recently been determined, which revealed important information on the action mode of VP-1620,21. As the structurally similar compounds of VP-16, a 4′-demethylepipodophyllotoxin (DMEP) derivative modified by a carbon-sulfur bond (i.e., 4β-S-(benzimidazole)-DMEP) (compound C-Bi) was developed in our previous work, which exhibited superior antitumor activity, the inhibition activity of target proteins Top2s, and apoptosis induction to that of VP-1622. However, the molecular mechanism underlying the difference is not well understood. Herein, the crystal structure of the catalytic core of hTop2β in complex with DNA and DMEP derivative was determined. The results can provide useful information for better understanding of the action mechanisms of Top2s-targeting antitumor agents.
Results
Detecting hTop2β-DNA covalent complexes using the ICE (in vivo complex of enzyme) bioassay
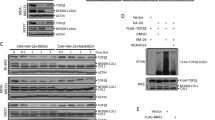
The ICE bioassay detected the hTop2β-DNA covalent complex formation in vivo with HeLa cells. Exponentially growing cultures were either treated with drugs (100 μM) or without drug. And then processed for covalent complexes. In the CsCl gradients (Fig. 1A), DNA mainly distributed at the expected density in fractions 9–12. Then, immunoblotting of the gradient fractions using an appropriate antibody measures hTop2β. As shown in Fig. 1B, free proteins were in the top fractions 1–8. No stabilized hTop2β-DNA intermediates were detected in the absence of drug, indicating a lack of cross-reactivity between hTop2β and DNA. In the presence of VP-16 or compound C-Bi, hTop2β was detected in the DNA peak fractions. It clearly suggested that the formation of hTop2β-DNA covalent complexes could be promoted under the action of VP-16 or compound C-Bi.
Detection of DMEP derivatives-induced hTop2β-DNA covalent complexes in HeLa cells. HeLa cells (1 × 107) were treated with 100 μM drugs for 30 min at 37 °C. (A) DNA profiles of untreated and 100 μM VP-16 or compound C-Bi treated cells. (B) Immunoblot data of gradient fractions from untreated and 100 μM VP-16 or compound C-Bi treated cells. The spots of compound C-Bi came from another nitrocellulose membrane (For complete blots we refer to Supplementary Fig. S1). The two membranes were operated and photographed under the same condition without overexposure or exposure adjustment.
DNA cleavage assay
To determine whether the DNA cleavage induced by compound C-Bi is associated with the interruption of topoisomearse II activity, a cell-free DNA cleavage assay using an enzyme-mediated negatively supercoiled pHOT1 plasmid DNA was applied. A representative gel image of the relaxation assay for determining the catalytic inhibition on hTop2β was presented in Fig. 2A. Topoisomerase IIβ poisons such as VP-16 were known to stabilize the topoisomerase IIβ-DNA complex that lead to DNA breaks and generate linear DNA23,24. In agarose gel, linear DNA (L) was difficult to enter and appeared at the top; whereas the relaxed DNA (R), and supercoiled DNA (S) move easily into the gel. As shown in Fig. 2A, compound C-Bi stabilized hTop2β cleavage complexes and exhibited the formation of linear DNA. This result shows that compound C-Bi acted as hTop2β poison. To more clearly describe the concentration-dependent effect on linear DNA, a gray-scale analysis was performed (Fig. 2B). It was observed that the DNA cleavage increased with an increase in the concentration of VP-16 and compound C-Bi. It indicated that VP-16 and compound C-Bi stabilized or trapped the Top2-cleaved DNA complex in a concentration-dependent manner.
Concentration dependent effect of compound C-Bi and VP-16 on the hTop2β-mediated cleavage of supercoiled pHOT1 plasmid DNA to produce linear DNA. (A) The linear DNA detection of different reaction systems. The concentration series were 0.1, 1, 5, 10, 20, 40, 80, 100 μM for compound C-Bi and VP-16. L, linear DNA; S, supercoiled DNA; R, relaxed DNA. (B) The gray scale value of the linear DNA amount. Data plotted are the mean ± SD of three separate experiments.
Binding of DMEP derivatives to hTop2β-DNA complex
To further investigate the binding affinity of compound C-Bi and VP-16 to hTop2β-DNA complex, surface plasmon resonance (SPR) was employed. As shown in Fig. 3, the response unit increased in a concentration-dependent manner. The equilibrium dissociation constant (KD) of compound C-Bi (45.9 ± 3.6 μM) were approximately 2.5 folds lower than that of VP-16 (113.6 ± 6.4 μM). These data indicated DMEP derivative exhibited higher affinity for hTop2β-DNA complex than VP-16.
The interaction between compound C-Bi and VP-16 with immobilized hTop2β-DNA complex was measured by surface plasmon resonance (SPR). (A) VP-16; (B) Compound C-Bi. In each case, the signal is proportional to the compounds concentration (For complete kinetics parameters we refer to Supplementary Table S1). The largest signal corresponds to the highest compound concentration and vice versa.
Structure basis of the complex
To provide insights into the interactions of compound C-Bi with hTop2β at the molecular level, we initially sought to investigate the molecular mechanism of action of compound C-Bi. Using the well-established system for hTop2β crystal growth20, the crystal of hTop2β cleavage complex containing two monomers of hTop2β and cleaved DNA chain was obtained under the action of compound C-Bi with the resolution of 2.54 Å (Table 1, Fig. 4A,B). The DNA was embedded in the hTop2β (Fig. 4C,D). All DNA base pairs were visible in the electron density maps (Supplementary Fig. S2). However, no molecule of compound C-Bi was observed in the stabilized hTop2β cleavage complex structures.
Structure of hTop2β-DNA complex. (A,B) Orthogonal views of the hTop2β-DNA complex from front view (A) and top view (B). (C,D) The cartoon representation of the hTop2β-DNA complex from front view (C) and top view (D). The dimeric hTop2β protein was shown as a surface representation and colored according to polypeptide chain. DNA was shown in yellow.
In order to understand the mechanism of compound C-Bi, the structures of hTop2β-DNA binary complex and hTop2β-DNA-etoposide ternary complex were compared. The RMSD value for this alignment was 3.604 Å (1304 to 1304 atoms). The structure of the hTop2β-DNA binary complex revealed a looser assembly pattern than in hTop2β-DNA-etoposide ternary complex (Fig. 5A,B) and other DNA-bound structures (Supplementary Table S2). However, the changes of structure were mainly located in the DNA-gate, and there was no obvious change in the C-gate. The distance between the main DNA-contacting domains of the two monomers was significantly increased under the action of compound C-Bi (Fig. 5C). The distance between drug-binding sites Arg503 of the two monomers was 26.62 Å in hTop2β-DNA-etoposide ternary complex and 34.54 Å in hTop2β-DNA binary complex, respectively. The distance between DNA-intercalating sites Ile872 of the two monomers was 52.65 Å in the ternary complex, and 58.71 Å in the binary complex.
Comparison the crystal structures of hTop2β-DNA complex (PDB: 5ZAD and 3QX3). (A,B) The superimposition of the hTop2β-DNA under the action of compound C-Bi (PDB ID 5ZAD: cyan and red) and hTop2β-DNA-VP-16 (PDB ID 3QX3: grey) structures from front view (A) and top view (B). VP-16 was colored blue. (C) Selected residues from the enclosed region were shown in an enlarged view to illustrate the change of distance between two monomers. The distances between residues involved in drug-binding (R503) and DNA-intercalating (I872) were indicated as dashed lines. Residues with labels from different structures were colored differently (3QX3 in green and black and 5ZAD in yellow and red). Oxygen and nitrogen atoms were colored red and blue, respectively. Labels belonging to the second monomer were flagged by a prime.
Arg503 is a major drug-contacting residue20,21. In hTop2β-DNA-etoposide ternary complex, for interacting with VP-16, the +5 guanine base could form a hydrogen bond with the main-chain carbonyl of Arg50320. In this work, perhaps because of the loose assembly of monomers, the distance between Arg503 and +5 guanine base was increased, and no hydrogen bond was observed (Fig. 6). Similarly, the phosphotyrosyl linkage that should existed between +1 nucleotide and active site residue Tyr821’ was weaken. DNA-intercalating isoleucine (Ile872) still bound between the +8 and +9 bases.
Detailed views of the DNA cleavage site of the hTop2β-DNA binary complex. Positive and negative numbers designated nucleotides downstream and upstream of the scissile phosphate. DNA was shown as blue sticks. Key residues were shown as yellow sticks. Oxygen and nitrogen atoms were colored red and blue, respectively. The two hTop2β monomers were colored differently. Labels belonging to the second monomer were flagged by a prime.
To compare the differences between DNA molecules, the superimposition of DNA molecules from hTop2β-DNA binary cleavage complex and hTop2β-DNA-etoposide ternary cleavage complex were shown. The insertion of VP-16 abolished the stacking interaction between the +1/+4 and −1/+5 base pairs. Thus, a drug-binding site for compound C-Bi between the same base pairs in hTop2β-DNA binary complex was assumed. More obvious twist have been observed in the DNA chains of binary cleavage complex (Fig. 7). Compared with that of VP-16, the DNA chain configuration was changed by approximately 40.3° of the angle and 3.9 Å of distance under the action of compound C-Bi.
The superimposition of DNA molecules from hTop2β-DNA binary cleavage complex (PDB ID 5ZAD: yellow) and hTop2β-DNA-etoposide ternary cleavage complex (PDB ID 3QX3: grey). VP-16 was shown in blue stick representation. The red arrow represented the conjectural binding site of compound C-Bi. The comparison of the changes in configuration by calculating the angle and the distance for the structure of the enclosed region were shown from close-up view. The angle calculation was based on the overlapping point of the top view projection of DNA chains. This overlapping point was approximately +6 base of hTop2β-DNA-etoposide ternary cleavage complex. The labels of base were colored orange (PDB ID 5ZAD) and black (PDB ID 3QX3), respectively.
Discussion
VP-16 is one the most effective anticancer drugs in clinical use, the mechanism of which was reported as stabilized the hTop2β-DNA complex by bonding at the two scissile bonds of a hTop2β-DNA cleavage complex. In our previous work, a novel 4β-sulfur-substituted DMEP derivative was developed, which exhibited superior antitumor activity to that of VP-1622. As the structurally similar compound of VP-16, the antitumor mechanism of compound C-Bi remains largely unknown.
Through ICE assays, DNA cleavage assay and kinetic analysis, we have identified compound C-Bi could bind to hTop2β, and stabilize the formation of ternary complex. To further understand the mechanism, the crystal structure (2.54 Å) of hTop2β cleavage complexes stabilized by compound C-Bi was determined. However, no molecule of compound C-Bi was observed in the hTop2β cleavage complex structures. In our study, the stable hTop2β-DNA complex cannot be formed without the action of DMEP derivative (data not shown). In the previous work, the hTop2β-DNA binary complex can only be obtained through soaking out of VP-16 from the hTop2βcore-DNA-etoposide ternary complex21. By comparison of the structure of hTop2βcore-DNA-VP-16 ternary complex (PDB: 3QX3) and hTop2β-DNA binary complex (PDB: 4J3N), the difference was only the absence of VP-16. It indicated that although there was no compound C-Bi in the complex structure, the crystal structure could still provide information for explaining the mechanism of compound C-Bi action.
To assess the possible mechanism of compound C-Bi, we compared the structure mode of hTop2βcore-DNA binary complex (PDB: 5ZAD) and hTop2core-DNA-etoposide ternary complex (PDB: 3QX3). In the crystal structure of hTop2βcore-DNA complex under the action of compound C-Bi, with the two monomers moving away from each other, the distances between the drug-contacting sites (Arg503 and Arg503′) and between the two DNA-intercalating isoleucines (Ile872 and Ile872′) are longer than those observed in VP-16-stabilized structures (Fig. 5, Supplementary Table S2). Increased spacing may suggest an intermediate state between a closed state25 and the open conformation26. It indicated the action of compound C-Bi needed a looser packing between the two monomers. It was speculated that the amplitude of hTop2β motions increased under the effect of compound C-Bi. Therefore, the intermediate states of the movement of two monomers could be easily captured.
The change of cleavage DNA was also observed with more significant twist of DNA chains (Fig. 7). Compared with VP-16, the action of compound C-Bi led to further migration of the two DNA chains. That may be caused by the increased distance between the two monomers. The interaction between the residues and the base was weakened or eliminated by the migration of the monomers. DNA chains were more significantly twisted, and some bases could not be paired with each other (+1/+4). Lack of base-pairing and torsion of the DNA chains leads to an inability to re-ligate the DNA break, thus preventing repair of the DNA damage. When Top2s play roles in replication, transcription, chromosome condensation, and segregation, Top2 alters DNA topology via the formation of a Top2-DNA cleavage complex26. Top2 poisons (e.g. VP-16) inhibit the religation of cleaved DNA resulting in DNA damage27. Accumulation of Top2-mediated DNA damage leads to cell death. It suggested that compound C-Bi played antitumor roles through increasing spacing of hTop2β monomers. The changes in hTop2β structure further caused double changes in the torsional direction and migration distance of the DNA chains, resulting in impeding religation of DNA.
In summary, to understand the structural basis of compound C-Bi action, we determined the interaction of compound C-Bi with hTop2β and DNA using X-ray crystallography. By comparing the structures of the complexes, the distance between the two monomers of hTop2β was increased, and significant twist of DNA chains was found under the action of compound C-Bi. It suggested that compound C-Bi played antitumor roles through increasing the change of hTop2β monomers structure and structural changes will further generated DNA damage. The structure information reported here advanced the understanding of the inhibitory mechanisms of Top2-targeting anticancer compounds.
Methods
Construction and expression of hTop2βcore
The hTop2βcore-PET51b plasmid was kindly provided by Dr. Nei-Li Chan at National Taiwan University20. The plasmid was transformed into Escherichia coli BL21 (DE3) pLysS cells. For expression, the transformed strain was grown in LB medium at 37 °C to an OD600 = 1.0. Isopropyl-β-D-thiogalactopyranoside (IPTG) (Biosharp, Seoul, Korea) was added to a final concentration of 0.3 mM, and protein expression was induced at 20 °C for 16 h.
hTop2βcore purification
The cells were centrifuged at 5000 rpm for 15 min (4 °C), and the pellets were resuspended in lysis buffer containing 50 mM NaPi (pH 7.4), 10% glycerol, 500 mM NaCl, 5 mM β-mercaptoethanol, 0.5 mM phenylmethanesul fonylfluoride, and 10 mM imidazole20. And then the cells were disrupted using a high pressure cell crusher (AH-1500, ATS, Canada) at 4 °C. Unbroken cells were removed by centrifugation at 15,000 rpm for 30 min. The hTop2βcore protein was isolated from the cell lysate supernatant using Ni-NTA column (Clontech, USA) and eluted with elution buffer containing 250 mM imidazole without NaCl. The resulting protein was loaded onto a HiPrep 16/10 Heparin FF column (GE Healthcare). The protein was eluted in a linear gradient over 10 column volumes with buffer A (30 mM Tris-HCl pH 7.5, 15 mM NaCl, 2 mM β-mercaptoethanol, and 1 mM EDTA) and buffer B (buffer A containing 1 M NaCl). The eluted fractions were pooled and purified on a gel filtration column (Superdex 200, GE Healthcare) in buffer C (buffer A containing 70 mM NaCl). The hTop2βcore protein (with molecular weight about 180 kDa) was collected and concentrated to 6.5 mg/ml for crystallization.
DNA substrate for crystallography
The DNA sequence 5′-AGCCGAGCTGCAGCTCGGCT-3′ of the double-stranded DNA substrate was prepared as previously described20. Synthesis and purification of DNA were performed by a commercial company (GenScript, Nanjing, China). The oligonucleotides were dissolved in buffer containing 30 mM Tris-HCl (pH 7.5), 70 mM NaCl, 2 mM β-mercaptoethanol, and 1 mM EDTA) and annealed at 55 °C to generate double-stranded DNA for crystallization.
ICE assay
The ICE bioassay was performed as previously described28,29. HeLa cells were cultured in 25 cm cell culture dishes at 37 °C, and a single Petri dish was used per treatment (90% confluent, approx 1 × 107 cells). The medium was removed and replaced with serum-free medium (1 mL) containing topoisomerase inhibitors (e.g., 100 μM etoposide or compound C-Bi, final concentration) and cells were incubated at 37 °C for 60 min. The negative control containing no drug (solvent only) was prepared in parallel. Then the drug containing medium was removed, and cells were lysed by the 1% sarkosyl equilibrated to 37 °C (2 mL/plate). The sarkosyl was squirted on the plate two to three times to ensure complete and rapid lysis. Cell lysates were layered onto a cesium chloride gradient in polyallomer tubes. The CsCl density gradients contained four different density steps (1.72, 1.49, 1.02, 0.84 g/mL). To prepare the gradients, a stock solution of CsCl (density = 1.86 g/mL) was diluted with 1 × TE (10 mM Tris-HCl, pH 7.5, 1 mM EDTA). The tubes were centrifuged in a Beckman SW41 rotor at 125,000 × g for 20 h at 20 °C. Fractions (0.2 mL) were collected from the top of the gradients. The DNA concentration was determined using a microplate reader at 260 nm (EON, BioTek, USA).
All fractions were diluted with the equivalent volume of 25 mM sodium phosphate buffer (pH 6.5) for 15 min, and blotted on a nitrocellulose membrane. After soaking the membrane in TBST (10 mM Tris-HCl, pH 7.5, 150 mM NaCl) for 15 min and blocking with TBST containing 5% nonfat dried milk for 2 h, the complexes of hTop2β-DNA and free hTop2β were detected using specific antibody probe (Proteintech Group, USA) at a 1:10000 dilution. The IRDye 800CW Goat anti-rabbit (LI-COR Biosystems, USA) (1:15000) was used as secondary antibody. The fluorescence signals were analyzed using the Odyssey imaging system (LI-COR Biosystems, USA).
DNA cleavage assay
Effect of compound C-Bi on hTop2β was measured using a topoisomerase II drug screening kit (TopoGEN, USA). The cleavage reactions were carried out in a final volume of 20 μL, containing 100 ng of supercoiled pHOT1 plasmid DNA, 3 units of hTop2β, 0–100 μM compound C-Bi (dissolved in 1% DMSO), and complete assay buffer (5×). After incubation at 37 °C for 30 min, the reaction was terminated by the addition of 2 μL sodium dodecyl sulphate (10%). Then the reaction mixtures were incubated with 2 μL of proteinase K (50 μg/mL) at 37 °C for 15 min to digest hTop2β. Final samples were mixed with 2 μL loading buffer (10×) and subjected to electrophoresis in 1% agarose.
Surface plasmon resonance
The SPR assay was performed at 25 °C using a BI2000 SPR system (BioSensing Instrument, USA). Before determination, 1 × cysteamine hydrochloride (Sigma-Aldrich) was covered onto the sensor chip in a dark. After 12 h incubation, the chip was further covered by the mixture solution of dextran sodium salt, N-hydroxysuccinimide and N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (All from Sigma-Aldrich) for 3 h. hTop2β and DNA (in a 1.2-fold molar ratio to protein) were incubated at 4 °C for 1 h, and then the hTop2β-DNA binary complex was immobilized onto the sensor chip. The running buffer was PBS (pH 6.5) containing 5% DMSO. 1 M ethanolamine hydrochloride solution (30 μL/min, 100 μL) was then injected to block the unreacted ester group. After that, compound C-Bi was injected to flow across the chip surface with the concentrations from 50 to 800 μM at a flow rate of 60 μL/min. Each analyte was in triplicate determination. Between measurements, a regeneration cycle was completed by applying 20 mM NaOH over the sensor chip to dissociate the compound. The kinetic parameters provided estimates of both association and dissociation rate constants (ka and kd), and from these values the equilibrium parameter (KD) for evaluating binding affinity of hTop2β-DNA for DMEP derivative was calculated using the relationship KD = kd/ka.
Crystallization
The protein sample was mixed with 2 mM compound C-Bi (in DMSO) and DNA substrate (in a 1.2-fold molar ratio to protein). Initial crystallization trials for the hTop2βcore-DNA-compound C-Bi complex were performed as previously reported20. 1 μL of concentrated hTop2βcore-DNA- compound C-Bi was mixed with an equal amount of reservoir solution and equilibrated against 500 μL of reservoir solution at 4 °C. The complex was crystallized by the hanging-drop vapor diffusion method using 100 mM magnesium chloride, 50 mM sodium citrate (pH 5.3), and 20% 2-methyl-2,4-pentanediol (MPD) as the precipitating agent, and 0.05% ethyl acetate (Hampton Research, USA) was used as additive. Crystals reached the maximum dimensions in around two weeks. Crystals were harvested by transferring into mother liquor containing 30% MPD and 1 mM corresponding compound before looping and flash-freezing in liquid nitrogen for data collection.
Structure determination
The X-ray diffraction data on the hTop2βcore-DNA complex were collected at the Shanghai Synchrotron Radiation Facility (SSRF) (Shanghai, China) beamline BL18U and BL19U130 and were processed using the HKL3000 program suite. The structure was solved by molecular replacement with the PHENIX MR (using the structure of hTop2βcore-DNA-VP-16 (PDB: 3QX3)20 as the search model). Then the structure was built and refined by Coot31 and PHENIX32 and analyzed by PyMOL33.
Accession code
The crystal structure of the complex of hTop2βcore-DNA, has been deposited with the RCSB Protein Data Bank under the accession code 5ZAD.
References
Champoux, J. J. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 70, 369–413 (2001).
Ju, B. G. et al. A topoisomerase IIβ-mediated dsDNA break required for regulated transcription. Science 312, 1798–1802 (2006).
Deweese, J. E. & Osheroff, N. The DNA cleavage reaction of topoisomerase II: wolf in sheep’s clothing. Nucleic Acids Res. 37, 738–748 (2009).
Chen, S. H., Chan, N. L. & Hsieh, T. S. New mechanistic and functional insights into DNA topoisomerases. Annu. Rev. Biochem. 82, 139–170 (2013).
Linka, R. M. et al. C-terminal regions of topoisomerase II alpha and II beta determine isoform-specific functioning of the enzymes in vivo. Nucleic Acids Res. 35, 3810–3822 (2007).
Mondal, N. & Parvin, J. D. DNA topoisomerase IIα is required for RNA polymerase II transcription on chromatin templates. Nature 413, 435–438 (2001).
Ye, J. et al. TRF2 and apollo cooperate with topoisomerase 2α to protect human telomeres from replicative damage. Cell 142, 230–242 (2010).
Yang, X., Li, W., Prescott, E. D., Burden, S. J. & Wang, J. C. DNA topoisomerase IIβ and neural development. Science 287, 131–134 (2000).
Lyu, Y. L. et al. Role of topoisomerase IIβ in the expression of developmentally regulated genes. Mol. Cell. Biol. 26, 7929–7941 (2006).
Baldwin, E. L. & Osheroff, N. Etoposide, topoisomerase II and cancer. Curr. Med. Chem. 5, 363–372 (2005).
Nitiss, J. L. Targeting DNA topoisomerase II in cancer chemotherapy. Nat. Rev. Cancer 9, 338–350 (2009).
Mcclendon, A. K. & Osheroff, N. DNA topoisomerase II, genotoxicity, and cancer. Mutat. Res-Rev. Mutat. 623, 83–97 (2007).
Lyu, Y. L. et al. Topoisomerase II beta mediated DNA double-strand breaks: implications in doxorubicin cardiotoxicity and prevention by dexrazoxane. Cancer Res. 67, 8839–8846 (2007).
Lee, C. H. et al. Anthracenedione-methionine conjugates are novel topoisomerase II-targeting anticancer agents with favorable drug resistance profiles. Biochem. Pharmacol. 83, 1208–1216 (2012).
Bax, B. D. et al. Type IIA topoisomerase inhibition by a new class of antibacterial agents. Nature 466, 935–940 (2010).
Mariani, A. et al. Differential targeting of human topoisomerase II isoforms with small molecules. J. Med. Chem. 58, 4851–4856 (2015).
Wohlkonig, A. et al. Structural basis of quinolone inhibition of type IIA topoisomerases and target-mediated resistance. Nat. Struct. Mol. Biol. 17, 1152–1163 (2010).
Leone, S., Basso, E., Polticelli, F. & Cozzi, R. Resveratrol acts as a topoisomerase II poison in human glioma cells. Int. J. Cancer 131, 173–178 (2012).
Li, T. K. & Liu, L. F. Tumor cell death induced by topoisomerase-targeting drugs. Annu. Rev. Pharmacol. Toxicol. 41, 53–77 (2001).
Wu, C. C. et al. Structural basis of type II topoisomerase inhibition by the anticancer drug etoposide. Science 333, 459–462 (2011).
Wu, C. C., Li, Y. C., Wang, Y. R., Li, T. K. & Chan, N. L. On the structural basis and design guidelines for type II topoisomerase-targeting anticancer drugs. Nucleic Acids Res. 41, 10630–10640 (2013).
Li, J. L. et al. Comparison of carbon-sulfur and carbon-amine bond in therapeutic drug: 4β-S-aromatic heterocyclic podophyllum derivatives display antitumor activity. Sci. Rep. 5, 14814 (2015).
Baviskar, A. T. et al. Switch in site of inhibition: a strategy for structure-based discovery of human topoisomerase IIα catalytic inhibitors. ACS. Med. Chem. Lett. 6, 481–485 (2015).
Vann, K. R. et al. Inhibition of human DNA topoisomerase IIα by two novel ellipticine derivatives. Bioorg. Med. Chem. Lett. 26, 1809–1812 (2016).
Schmidt, B. H., Burgin, A. B., Deweese, J. E., Osheroff, N. & Berger, J. M. A novel and unified two-metal mechanism for DNA cleavage by type II and IA topoisomerases. Nature 465, 641–645 (2010).
Berger, J. M., Gamblin, S. J., Harrison, S. C. & Wang, J. C. Structure and mechanism of DNA topoisomerase II. Nature 379, 225–232 (1996).
Pommier, Y., Leo, E., Zhang, H. & Marchand, C. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem. Biol. 17, 421–433 (2010).
Subramanian, D., Furbee, C. S. & Muller, M. T. ICE bioassay: isolating in vivo complexes of enzyme to DNA. Mol. Biol. 95, 137–147 (2001).
Subramanian, D., Kraut, E., Staubus, A., Young, D. C. & Muller, M. T. Analysis of topoisomerase I/DNA complexes in patients administered topotecan. Cancer Res. 55, 2097–2103 (1995).
Wang, Q. S. et al. The macromolecular crystallography beamline of SSRF. Nucl. Sci. Tech. 26, 10102 (2015).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Schrodinger, L. L. C. The PyMOL molecular graphics system. Version 1, 3r1 (2010).
Acknowledgements
We thank Huan Zhou at Shanghai Synchrotron Radiation Facility (SSRF) beamline BL17U for their on-site assistance. We thank the research associates, Delin Zhang, at the Center for Protein Research (CPR), Huazhong Agricultural University, for his technical support. Financial supports from the National Natural Science Foundation for Distinguished Young Scholars (Grant No. 21625602), the National Natural Science Foundation of China (Grant Nos 21838002, 81503112, 21506049, and 31570054), and Hubei Provincial Science and Technology Innovation Major Project (2017ACA173) are gratefully acknowledged.
Author information
Authors and Affiliations
Contributions
Y.J.T. conceived the project. L.W.Z. designed the experiments; L.Y.S. implemented the analysis work flow and conducted the experiments. L.Y.S. and L.W.Z. contributed to the analysis with constructive discussions, L.Y.S. prepared all figures and tables, L.Y.S., L.W.Z. and Y.J.T. wrote the manuscript. All authors reviewed, commented on, and approved the final manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sun, LY., Zhu, LW. & Tang, YJ. Increasing the distance between two monomers of topoisomerase IIβ under the action of antitumor agent 4β-sulfur-(benzimidazole) 4′-demethylepipodophyllotoxin. Sci Rep 8, 14949 (2018). https://doi.org/10.1038/s41598-018-33366-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-33366-2
Keywords
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.