Abstract
Only three immediate early genes (IE) BICP0, BICP4 and BICP22 of Bovine herpesvirus 1 (BoHV-1) are known. These genes are expressed coordinately and their promoters are well characterized. We provide evidence for expression of three additional IE genes of BoHV-1 i.e. UL21, UL33 and UL34. These genes are expressed in the presence of cycloheximide (CH) at the same time as known IE genes. Surprisingly, the promoters of newly identified IE genes (UL21, UL33, UL34) lack the OCT-1 binding site, a considered site of transactivation of the BoHV-1 IE genes. The other difference in the promoters of the newly identified IE genes is the presence of TATA box at near optimal site. However, all the IE genes have similar spatial placements of C/EBPα, DPE and INR elements.
Similar content being viewed by others
Introduction
Bovine herpesvirus 1 (BoHV-1) primarily infects cattle and is associated with respiratory and reproductive venereal diseases along with other clinical symptoms like conjunctivitis, abortion, encephalitis, enteritis, and generalized disease in newborn calves1. BoHV-1 is a member of the subfamily Alphaherpesvirinae which includes Human Herpesvirus 1 (HHV-1), the prototype virus for the Alphaherpesvirinae subfamily. BoHV-1 has a class D herpesvirus genome. Its genome contains two unique regions, the unique long (UL) and the unique short (US). US region is flanked by inverted repeats (IR) and terminal repeats (TR)2. Like HHV-1, viral gene expression in BoHV-1 is regulated temporally in three distinct phases: alpha or immediate-early (IE), beta or early (E) and gamma or late (L)3,4,5. Expression of IE genes initiates from 1 hour post infection (p.i.) and maximizes between 3 to 4 hour p.i6.
The IE genes are important for the replication of BoHV-1 as they are required for activation of E and L genes and also for subsequent down regulation of IE genes themselves5. During its lytic cycle, BoHV-1 produces three major IE transcripts namely IE4.2 encoding BoHV-1 infective cell protein 4 (BICP4); IE2.9 encoding BICP0 and IE1.7 encoding BICP224. These BICP genes are counterparts to infected cell proteins (ICP) 0, 4 and 22 of HHV-17. The IE transcripts are grouped under two divergent IE transcription unit (IETU); IE4.2 and IE2.9 belongs to IETU1 and IE1.7 comes under IETU24,5,8 but recently it has been shown that BICP0, BICP4 and BICP22 are transcribed from three independent transcription units9.
In HHV-1 infected cells, viral gene expression is initiated when the IE genes are transactivated by α gene trans-inducing factor α-TIF, also known as VP16 or Vwm6510,11 but α-TIF does not directly bind DNA, rather it forms a complex with the octamer binding protein (OCT-1) which recognizes and binds to the DNA motifs with the consensus sequence TAATGARAT (OCT-1 motif)10,12 located in the promoters of the HHV-1 IE genes. This transcriptional program has been well documented in HHV-1, but other members of Alphaherpesvirinae seems to follow somewhat similar transcriptional pattern12,13,14,15. The importance of OCT-1 can be inferred from the fact that it is essential for the sequence specific action of α-TIF16.α-TIF provides DNA-binding specificity for formation of the α-TIF-OCT-1 complex because it associates with OCT-1 protein only at the OCT-1-binding motif17. The binding of OCT-1 protein with α-TIF brings the complex into the proximity of the transcription start sites18,19,20,21. Katan et al. have shown that deletion in POU domain of OCT-1 protein affects complex formation with α-TIF22.
Many α-TIF homologs with similar IE promoter activation properties have been identified in herpes viruses. For example, α-TIF homologs from BoHV-1, varicella-zoster virus, and Equine herpesvirus 1 transactivates IE promoters and can form α-TIF-OCT-1 containing regulatory complexes on OCT-1 binding motifs12,17,23,24. The homolog of tegument protein α-TIF in BoHV-1 is b-transinducing factor (bTIF) and it is responsible for initiating IE viral gene expression in a similar fashion to HHV-1 α-TIF13,25.
There is a report which indicates that OCT-1 protein is not the only cellular factor that can transactivate BoHV-1 gene expression25. The study suggested the CCAAT enhancer-binding protein alpha (C/EBPα) has the ability to transactivate IE genes. C/EBPs are a family of transcription factors that contains a conserved, basic- leucine zipper domain involved in DNA binding26 and belongs to the family of leucine zipper class DNA binding protein27.
The transcriptional regulation and computational studies of BoHV-1 in the past have been focused on the three major IE genes3,4,5,8. A random insertional mutagenesis study by Robinson et al. mentioned seven genes of unknown regulatory control28. This study aimed to ascertain the temporal expression patterns of three BoHV-1 genes viz. UL21, UL33 and UL34 and also evaluated their possible transcriptional regulatory elements.
Results
The IE nature of UL21, UL33 and UL34 genes
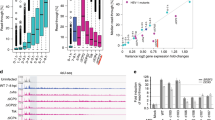
To assess if UL21, UL33 and UL34 genes are IE genes, the expression of these genes was studied in BoHV-1 infected cells in the presence and absence of cycloheximide (CH). The transcription of the known IE gene, BICP0 as a control was not inhibited by CH (Fig. 1E) while CH effectively inhibited the expression of early gene thymidine kinase (TK) and late gene glycoprotein B (gB) without affecting the expression of IE genes (Fig. 1D). Figure 1A–C shows the amplification of UL21, UL33, UL34. BICP0, TK and gB genes at different time point in BoHV-1 infected cells in the presence and absence of CH. The sequence of the PCR amplified product was identical to that of the targeted template region (data not shown)3,6,29. The GAPDH control confirmed the quantity of the total RNA templates (Supplementary Fig. 1G).
(A) Agarose gel Electrophoresis of UL21 PCR amplicon (234 bp) at time points after cycloheximide treatment of BoHV-1 infected MDBK cells. Lane M: 100bp DNA marker (Thermo scientific). Lane 1: 1 hours. Lane 2: 2 hours. Lane 3: 3hours. Lane 4: 4 hours. Lane 5: 5 hours. (B) Agarose gel Electrophoresis of UL34 PCR amplicon (253 bp) at time points after Cycloheximide treatment of BoHV-1 infected MDBK cells. Lane 1: 5 hours. Lane 2: 4 hours. Lane 3: 3 hours. Lane 4: 2 hours. Lane 5: 1 hours. Lane M: 100bp DNA marker (Thermo scientific). (C) Agarose gel Electrophoresis of UL33 PCR amplicon (211 bp) at time points after Cycloheximide treatment of BoHV-1 infected MDBK cells. Lane M: 100bp DNA marker (Thermo scientific). Lane 1: 1 hours. Lane 2: 2 hours. Lane 3: 3 hours. Lane 4: 4 hours. Lane 5: 5 hours. Lane 6: healthy MDBK used as negative control. (D) Agarose gel Electrophoresis of gB PCR amplicon (468 bp) at time points with and without Cycloheximide treatment of BoHV-1 infected MDBK cells in late hours. Lane M: 100bp DNA marker (Thermo scientific). Lane 1: No PCR product of gB gene after 12 hour post infection of BHV-1 without treatment of cycloheximide. Lane 2: No PCR product of gB gene after 12 hour post infection of BHV-1 with treatment of cycloheximide. Lane 3: PCR product of 468 bp of gB gene after 14 hour post infection of BHV-1 without treatment of cycloheximide. Lane 4: No PCR product of gB gene after 14 hour post infection of BHV-1 with treatment of cycloheximide. Lane 5: PCR product of 468bp of gB gene after 16 hour post infection of BHV-1 without treatment of cycloheximide. Lane 6: No PCR product of gB gene after 16 hour post infection of BHV-1 with treatment of cycloheximide. Lane 7: healthy MDBK control. Lane M: 1Kb DNA Ladder (Thermo Scientific). (E) Agarose gel Electrophoresis of BICP0 PCR amplicon (209 bp) at time points after Cycloheximide treatment of BoHV-1 infected MDBK cells. Lane 1: 5 hours. Lane 2: 4 hours. Lane 3: 3hours. Lane 4: 2 hours. Lane 5: 1 hours. Lane M: 100bp DNA Ladder (Thermo Scientific). (F) Agarose gel Electrophoresis of TK PCR amplicon (216 bp) at time points with and without Cycloheximide treatment of BoHV-1 infected MDBK cells in early hours. Lane M: 100bp DNA marker (Thermo scientific). Lane 1: PCR product of 216 bp of TK gene after 6 hour post infection of BHV-1 without treatment of cycloheximide. Lane 2: No PCR product of TK gene after 6 hour post infection of BHV-1 with treatment of cycloheximide. Lane 3: PCR product of 216 bp of TK gene after 8 hour post infection of BHV-1 without treatment of cycloheximide. Lane 4: No PCR product of TK gene after 8 hour post infection of BHV-1 with treatment of cycloheximide. Lane 5: healthy MDBK control.
The experiment was repeated three times and found similar results. Results indicate that UL21, UL33 and UL34 are IE genes like BICP0, BICP4 and BICP22 since their expression is not inhibited in presence of CH.
Promoters of UL21, UL33 and UL34 genes
To delineate the promoter regions of UL21, UL33 and UL34 genes, the transcription start sites (TSS) of these genes were first identified by 5′-RLM-RACE. Each gene gave one amplification product differing in size in RLM-RACE (Fig. 2A–C) suggesting one TSS for each gene. The BICP0, BICP4 and BICP22 also have one transcription start site9. The distance between TSS and putative translation start site (ATG) varied among these three genes. This distance was 21 bp in UL21, 127 bp in UL33 and 48 bp in UL34.
(A) Agarose gel Electrophoresis showing RACE amplified products of UL33 gene Lane M: 100bp DNA marker (Thermo scientific). Lane 1: PCR product of 300bp of UL33gene. (B) Agarose gel Electrophoresis showing RACE amplified products of UL21 gene. Lane M: 100bp DNA marker (Thermo scientific). Lane 1: PCR product of 250bp of UL21 gene. (C) Agarose gel Electrophoresis showing RACE amplified products of UL34 gene Lane M: 100bp DNA marker (Thermo scientific). Lane 1: PCR product of 300bp of UL34 gene.
Comparative computational analysis of UL21, UL33 and UL34 genes with BICP0, BICP4 and BICP22
The comparative computational analysis between the putative promoters of UL21, UL33, UL34 and BICP0, BICP4, BICP22 genes revealed some interesting findings. For computational analysis, we took 1000 bp upstream and 100 bp downstream of the identified TSS sites. The UL21, UL33 and UL34 group showed presence of TATA boxes at optimal position30,31,32 (−30, −28 and −25 bp respectively) from the TSS as shown in Fig. 3 while the TATA box was absent in BICP4, BICP0 and BICP22 at appropriate position though TATA box like sequence was observed at position far upstream from the TSS (−251, −143 and −359 bp respectively). The OCT-1 like sequences were absent in the UL group of IE genes despite intensive examination with relaxed parameters keeping three base mismatch. The OCT-1 motifs were identified in promotor regions of the three known IE genes (Fig. 3). INR (Initiator elements) and DPE (Downstream Processing elements) are important regions in core promoters required for RNA polymerase binding33,34,35. Their presence aids in the prediction of probable promoter sites. INR and DPE were present in both the groups of IE genes and their spatial positioning was almost similar. The optimum position of DPE is at +28 to +32 from the TSS32,34. The maximum variation from the optimum position observed was 10 bp.
In BoHV-1, report shows that C/EBPα has the ability to transactivate IE genes25. Searching for C/EBPα binding sequences showed conserved C/EBPα motifs in all IE gene (UL21, UL33, UL34, BICP0, BICP4 and BICP22) promoters at similar spatial position relative to the TSS. The nearest conserved position of C/EBPα relative to the TSS was −685, −678, −646, −685, −647, −785 bp in BICP4, BICP0, BICP22, UL21, UL33 and UL34 respectively (Fig. 3).
Comparative expression of basal promoters of UL 21and BICP0 genes
The UL group of IE genes contains TATA box in optimal positions30 while TATA boxes identified in promoter regions of the BICP group were not at the optimal position suggesting that they have no role in RNA polymerase binding. To test if TATA box in optimal position influences basal promoter activity of UL and BICP promoters, the promoter region 300 bp upstream from TSS of one representative from each group (UL21 TATA box at −30 and BICP0 TATA box at −148 base upstream) was cloned in promoterless pDSred Express 2–1 vector and transfected into MDBK cells. The expression of reporter gene was measured in flow cytometer (Fig. 4). The data shows higher expression of UL21 promoter as compared to BICP0 promoter. However, this was not reflected in the temporal expression by real time PCR. Real Time PCR showed almost similar expression of both UL and BICP group of IE genes (Fig. 5). The expression study results suggest that the factors upstream of 300 bp may influence expression in vivo. Flow cytometry (translation) and real time PCR (transcription) data reflect two different parameters which are not necessarily comparable. Since, we have used same reporter gene having identical sequence, it is assumed that they will have same degradation, stability, translation efficiency etc. and so may reflect mRNA copy number.
Discussion
Presence of an OCT-1 motif and its other possible variants is an important feature in all the IE genes in HHV-1 identified to date. It is assumed that binding of OCT-1 by bTIF helps in the recruitment of this protein complex to the OCT-1 motif (IFTATGRAAT sequence motif) to initiate the transcription of IE genes in the BoHV-1 infection process22. This is the standard model for the beginning of transcription in HHV-1 and many other herpesviruses as well4,5,10,12,14,36. There are reports showing that C/EBPα can transactivate IE genes of BoHV-125,37,38,39, in vitro and also that bTIF can activate promoters that lack OCT-1 motifs provided an alternate mechanism for DNA binding is available10. We recently reanalyzed the promoters of known IE gene of BoHV-19 and observed that instead of two4,5,40, there are three IE transcription units and each of the transcription units have similarly placed INR, DPE and OCT-1 motifs thus reinforcing the hypothesis of OCT-1 having a role in the transactivation of IE genes in BoHV-1.
In this study, we identified three BoHV-1 IE genes that lack OCT-1 site but have a similarly located C/EBPα site as present in known IE genes. Since C/EBPα has the ability to transactivate genes, it is possible that the newly identified IE genes are getting transactivated through it25. This is a reasonable assumption which would need further experimentation to prove it. The second point to consider is the functional role if any for these newly identified IE genes. At present, the expression of these genes in virus infection in vitro or in vivo is not known consequently no functional role is ascribed to them. Putatively, they code for late proteins (UL21: tegument protein, UL3: DNA packaging, UL34: nuclear egress protein)28,41. Based on this information, it is difficult to make any guess on their functional role at present.
Identifying gene expression cascade is important in virus multiplication and infection and can potentially be used for identifying targets for intervention. A new gene expression cascade has been identified in this study. Further work is needed to explore the importance of these IE genes in BoHV-1 and possibly other herpesviruses.
Methods
Cell Culture and virus infection
Madin-Darby Bovine Kidney (MDBK) cells were obtained from the National Centre for Cell Sciences (NCCS), Pune, India. The cells were cultured and maintained in DMEM (high glucose) (HyClone) supplemented with 10% FBS (HyClone), penicillin 100 U/mL, streptomycin 100 μg/mL at 37 °C and 5% CO2 in 25 cm2 -flasks (Nunc). An Indian isolate of BoHV-1 Virus strain (BoHV1/IBR 216 II/ 1976/India) maintained at Immunochemistry laboratory, Division of Biochemistry, IVRI, Izatnagar was used for the study.
Cycloheximide treatment
Sub-confluent (60–70% confluency) MDBK (maintained at Immunochemistry laboratory, Biochemistry Division of IVRI) monolayer in 12 well culture plate were infected with BoHV-1 at 10 multiplicity of infection (moi) along with CH at a concentration of 100 µg/mL and incubated at 37 °C until harvesting of RNA at different time intervals (1, 2, 3, 4, 5 hr.). CH was used to identify IE transcripts and added as described by Misra et al.3 and Wirth et al.3,8. BoHV-1 infected MDBK cells (without CH) were used as control. To check the efficiency of CH treatment, known early and late genes of BoHV-1 were used as controls in PCR. RNA was harvested at 6, 8, 10 and 12, 14, 16 hours post infection for TK for early gene and gB for late gene marker respectively. GAPDH primer pair (GAPDH/F and GAPDH/R as mentioned in Table 1), was used as an internal control for RNA quantity and amplification efficiency.
RNA Isolation
RNA was extracted at different time intervals i.e., 1, 2, 3, 4 and 5 hour post infection. For extraction of total RNA, 1 mL TRIZOL (Life technologies) was added directly to the virus infected monolayer and mixed by pipetting. The rest of the RNA extraction was as per the manufacturer’s protocols. The RNA pellet recovered after centrifugation at 10,000 × g for 15 minutes was washed once with 70% ethanol and then resuspended in 20 µL of nuclease free water. Before cDNA preparation and PCR amplification, isolated total RNA was treated with DNase I (1µ/µL) (Thermo scientific) at 37 °C for 30 min, followed by enzyme inactivation with 50 mM EDTA (2 µL) at 65 °C for 10 min. Same method for RNA isolation was followed for 6, 8, 10 and 12, 14 and 16 hour post infection, DNase treated RNA was used as negative control in PCR.
Oligonucleotide primers
BoHV-1 complete genome (AJ004801) from NCBI GenBank was used as reference to design gene specific forward and reverse primers to amplify the 5′ regions of the UL21, UL33 and UL34 genes. For RACE, universal primers (outer and inner) were provided with the RLM-RACE kit to recognize the 5′ end of the cDNA. Two sets of outer and inner (nested) gene specific reverse primers were designed to amplify the part of UL21, UL33 and UL34 genes. All the primer sets for PCR reactions were designed using GeneTool Lite (Biotools Incorporated Ltd.). The primer sequences and other relevant details of all the primers used in this study are given in Tables 1 and 2.
5′-RLM-RACE for TSS identification
For RACE, RNA was isolated from BoHV-1 infected MDBK cells in 25 cm2 cell culture flask at two hours post infection. RACE was performed using 5′-RLM-RACE (RNA Ligase Mediated Rapid Amplification of cDNA Ends) kit (Ambion, Life Technologies) according to the manufacturer’s recommendations. In brief, 5 µL of cDNA was subjected to the first round of PCR amplification using the adaptor primer (outer) provided in the kit and gene-specific reverse primer. The reaction was carried out in a thermal cycler (Biometra, U.K.). The cycling conditions were denaturation at 95 °C for 2 minutes, followed by 34 cycles of denaturation at 95 °C for 30 seconds, annealing touchdown (from 65 to 55 °C) for 45 seconds and extension at 72 °C for 1 minute, with a final extension at 72 °C for 5 minutes. After amplification, the reaction products were diluted 20-fold with water, and 3 µl DNA were then re-amplified for 30 cycles using the same reaction conditions with nested and adaptor primer (inner primer provided in the kit). Positive PCR products were eluted from 1% agarose gel and ligated into pGEM-TEasy vector (Promega) and sequenced through vendors (Bioserves).
Reverse transcription and Polymerase Chain Reaction (PCR)
cDNA was synthesized from DNase I treated RNAs using Revert Aid cDNA kit (Thermo Scientific). PCR was performed using the specific primers (Table 1) in 20 µL reaction mix. BICP0 (IE genes) was used as a control to confirm IE genes while TK and gB as a control for and L genes respectively. DNase I treated RNA was used as a control for genomic DNA contamination for each reaction. Each RT-PCR reaction contained the following: 10X Dream Taq buffer, 10pmol forward and reverse primers each, 1 µL DMSO, 1U Dream Taq polymerase (Thermo scientific) and 2 µL of cDNA. The reaction was carried out with an initial denaturation at 95 °C for 10 min followed by 34 cycle of denaturation at 95 °C for 30 Sec, annealing was done at 62 °C for 45 sec followed by extension at 72 °C for 1 min with a final extension at 72 °C for 5 min. The amplified PCR products were cloned and sequenced through vendors (Bioserves) to confirm specific amplification.
Monitoring the expression of UL21, UL33 and UL34 by real-time PCR
Real time PCR was performed for undefined genes (UL21, UL33 and UL34) along with known IE genes (BICP0, BICP4 and BICP22) with specific primers (Table 1). The data for BICP22 and UL34 were not included because of primer dimers despite several attempts were made to standardize it. Real Time PCR was performed to determine the expression pattern of undefined genes and to compare their expression pattern to known IE genes. Sub-confluent MDBK cells maintained in 12 well plate were infected with BoHV–1 virus at 10 m.o.i. Cells were harvested at different time intervals from 1, 2, 3, 4 and 5 hrs post infection (h.p.i). RNA extraction, DNase I treatment and cDNA synthesis was done as described in previous sections. The reaction was performed by SYBR green method using iCycler iQ™ Real-Time PCR Detection System (BIO-RAD). For each gene, 20 µL reaction mix was prepared in 0.2 ml PCR tube on ice in triplicates along with housekeeping gene Glyceraldehyde 3 phosphate dehydrogenase and non-template controls (NTC). The cycling conditions were 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 sec, 62 °C for 30 sec, 72 °C for 45 sec. The comparative CT (cycle threshold) method was employed for relative quantification of genes.
Construction of reporter plasmid constructs and Transient Transfection
Basal promoter regions of BICP0 and UL21 were PCR amplified and cloned into promoter less pDSRED Express 2-1 (Clonetech) vector.
All plasmids used for transfection were prepared with Sureprep Plasmid Endofree Maxi Kit (Genetix) according to manufacturer’s instructions. The recombinant plasmids were transfected into 70–80% confluent MDBK cells in a 24 well plate using PolyFect transfection reagent (QIAGEN, Germany) as per the manufacturer’s instructions. A total of 600 ng of recombinant plasmids were mixed with serum free Dulbecco’s Minimal Essential Medium (DMEM, optimum) and 3 µL of Polyfect reagent. All samples were kept in triplicates and incubated for 24 hours.
Flow Cytometry
Transfected cells were harvested and washed with prewarmed (37 °C) Phosphate Buffer Saline (PBS). Cells were dislodged with 50 µL of prewarmed Trypsin Versene Glucose solution (TVG) and pelleted by adding 300 µL PBS and centrifuged at 3,000 rpm for 5 min. The pellet was resuspended in 300 µL PBS and analyzed in FACScalibur (BD Dickinson, USA) in FL2 channel.
Computational identification of promoter elements
To analyze the putative promoter regions, 1000 nucleotide upstream and 100 nucleotide downstream of TSS was selected for searching the putative promoter core elements. Promoter regions were identified using the online web server PROMOTER SCAN (http://www-bimas.cit.nih.gov/molbio/proscan/) and the downstream core elements were identified using YAPP online webserver (http://www.gene-regulation.com/cgi-bin/pub/programs/patch/bin/patch.cgi). The promoter sequences were also checked with other softwares like TRANSFAC, BDGP-Neural Network Promoter Prediction.
References
Hulce, D. A. Identification of the immediate-early products of bovine herpesvirus-1. Retrospective Theses and Dissertations. Iowa Stae University (1990).
Muylkens, B., Thiry, J., Kirten, P., Schynts, F. & Thiry, E. Bovine herpesvirus 1 infection and infectious bovine rhinotracheitis. Vet. Res. 38, 181–209 (2007).
Misra, V., Blumenthal, R. M. & Babiuk, L. A. Proteins Specified by bovine herpesvirus 1 (infectious bovine rhinotracheitis virus). J. Virol. 40, 367–378 (1981).
Wirth, U., Vogt, B. & Schwyzer, M. The three major immediate-early transcripts of bovine herpesvirus 1 arise from two divergent and spliced transcription units. J. Virol. 65, 195–205 (1991).
Wirth, U. V. et al. Immediate-early RNA 2.9 and early RNA 2.6 of bovine herpesvirus 1 are 3′ coterminal and encode a putative zinc finger transactivator protein. J. Virol. 66, 2763–2772 (1992).
Honess, R. W. & Roizman, B. Regulation of herpesvirus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J. Virol. 14, 8–19 (1974).
Vogt, B., Schwyzer, M. & Ko, R. Immediate-early protein BICP22 of bovine herpesvirus 1 trans-represses viral promoters of different kinetic classes and is itself regulated by BICP0 at transcriptional and posttranscriptional levels. Arch. Virol. 1, 2447–2464 (1997).
Wirth, U. V., Gunkel, K., Engels, M. & Schwyzer, M. Spatial and temporal distribution of bovine herpesvirus 1 transcripts. J. Virol. 63, 4882–9 (1989).
Pokhriyal, M. et al. Bovine Herpes Virus 1 Major Immediate Early Transcription Unit 1 (IETU-1) Uses Alternative Promoters to Transcribe BICP0 and BICP4 Transcripts. Curr. Microbiol. 72, 420–5 (2016).
Misra, V., Walker, S., Hayes, S. & O’Hare, P. The bovine herpesvirus alpha gene trans-inducing factor activates transcription by mechanisms different from those of its herpes simplex virus type 1 counterpart VP16. J. Virol (1995).
Campbell, M. E. M., Palfreyman, J. W. & Preston, C. M. Identification of herpes simplex virus DNA sequences which encode a trans-acting polypeptide responsible for stimulation of immediate early transcription. J. Mol. Biol. 180, 1–19 (1984).
Misra, V., Bratanich, A. C., Carpenter, D. & O’Hare, P. Protein and DNA elements involved in transactivation of the promoter of the bovine herpesvirus (BHV) 1 IE-1 transcription unit by the BHV alpha gene trans-inducing factor. J. Virol. 68, 4898–909 (1994).
Workman, A. M. Regulation of Bovine Herpesvirus 1 (BHV-1) Productive Infection by Cellular Transcription Factors. 1 (2011).
Kristie, T. M., Lebowitz, J. H. & Sharp, P. A. The octamer-binding proteins form multi-protein -DNA complexes with the HSV aoTIF regulatory protein. EMBO J. 8, 4229–4238 (1989).
LaMarco, K. L. & McKnight, S. L. Purification of a set of cellular polypeptides that bind to the purine-rich cis-regulatory element of herpes simplex virus immediate early genes. Genes Dev. 3, 1372–1383 (1989).
Nicholl, M. J. & Preston, C. M. Inhibition of herpes simplex virus type 1 immediate-early gene expression by alpha interferon is not VP16 specific. J. Virol. 70, 6336–6339 (1996).
Huang, C. C. & Herr, W. Differential control of transcription by homologous homeodomain coregulators. Mol. Cell. Biol. 16, 2967–76 (1996).
Ingles, C. J., Shales, M., Cress, W. D., Triezenberg, S. J. & Greenblatt, J. Reduced binding of TFIID to transcriptionally compromised mutants of VP16. Nature 351, 588–90 (1991).
Klemm, R. D., Goodrich, J. A., Zhou, S. & Tjian, R. Molecular cloning and expression of the 32-kDa subunit of human TFIID reveals interactions with VP16 and TFIIB that mediate transcriptional activation. Proc Natl Acad Sci USA 92, 5788–5792 (1995).
Lin, Y. S., Ha, I., Maldonado, E., Reinberg, D. & Green, M. R. Binding of general transcription factor TFIIB to an acidic activating region. Nature 353, 569–71 (1991).
Uesugi, M. Induced Helix in the VP16 Activation Domain upon Binding to a Human TAF. Science (80-.). 277, 1310–1313 (1997).
Katan, M., Haigh, a., Verrijzer, C. P., van der Vliet, P. C. & O’Hare, P. Characterization of a cellular factor which interacts functionally with Oct-1 in the assembly of a multicomponent transcription complex. Nucleic Acids Res. 18, 6871–6880 (1990).
Moriuchi, H., Moriuchi, M., Straus, S. E. & Cohen, J. I. Varicella-zoster virus open reading frame 10 protein, the herpes simplex virus VP16 homolog, transactivates herpesvirus immediate-early gene promoters. J. Virol. 67, 2739–46 (1993).
Purewal, A. S. et al. Equid herpesviruses 1 and 4 encode functional homologs of the herpes simplex virus type 1 virion transactivator protein, VP16. Virology 198, 385–9 (1994).
Meyer, F. & Jones, C. The cellular transcription factor, CCAAT enhancer-binding protein alpha (C/EBP-alpha), has the potential to activate the bovine herpesvirus 1 immediate-early transcription unit 1 promoter. J. Neurovirol. 15, 123–30 (2009).
Ramji, D. P. & Foka, P. CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem. J. 365, 561–575 (2002).
Wedel, A. & Loms Ziegler-Heitbrock, H. W. The C/EBP family of Transcription factors. Immunobiology 193, 171–185 (1995).
Robinson, K. E., Meers, J., Gravel, J. L., McCarthy, F. M. & Mahony, T. J. The essential and non-essential genes of Bovine herpesvirus 1. J. Gen. Virol. 89, 2851–63 (2008).
Honess, R. W. & Roizman, B. Regulation of herpesvirus macromolecular synthesis: sequential transition of polypeptide synthesis requires functional viral polypeptides. Proc. Natl. Acad. Sci. USA 72, 1276–1280 (1975).
Burke, T. & Kadonaga, J. Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters. Genes & Dev. 10, 711–724 (1996).
Elizabeth, M. B. et al. Going the Distance: A current view of enhancer action. Science. 281, 5373–60 (1998).
Michael, H. Molecular genetics of the RNA Polymerase II general transcriptional machinery. Microbiology and Molecular Biology Reviews. 62, 465–503 (1998).
Roeder, R. The role of general initiation factors in transcription by RNA Polymerase II. Trends Biochem.Sci. 21, 327–335 (1996).
Smale, S. T. Core promoter architecture for eukaryotic protein-coding genes, p. 63–80.In R. C. Conaway and J. W. Conaway (ed.), Transcription: mechanisms and regulation. Raven Press, Ltd., New York, N.Y (1994).
Smale, S. T. & Baltimore, D. The “initiator” as a transcription control element. Cell. 57, 103–113 (1989).
Spector, D., Purves, F. & Roizman, B. Role of alpha-transinducing factor (VP16) in the induction of alpha genes within the context of viral genomes. J. Virol. 65, 3504–3513 (1991).
Meyer, F. et al. A protein encoded by the bovine herpesvirus 1 (BHV-1) latency related gene interacts with specific cellular regulatory proteins, including the CCAAT enhancer binding protein alpha (C/EBP-a). J Virol. 81(59), 67 (2007).
Meyer, F. et al. Identification of a novel protein encoded by the latency-related gene of bovine herpesvirus 1. J Neurovirology. 13, 569–578 (2007).
Workman, A., Perez, S., Doster, A. & Jones, C. Dexamethasone Treatment of Calves Latently Infected with BovineHerpesvirus 1 Leads to Activation of the bICP0 Early Promoter, in Partbythe Cellular Transcription Factor C/EBP-Alpha. J Virol. 83(17), 8800–8809 (2009).
Schwyzer, M., Wirth, U. V., Vogt, B. & Fraefel, C. BICP22 of bovine herpesvirus 1 is encoded by a spliced 1.7 kb RNA which exhibits immediate early and late transcription kinetics. J. Gen. Virol. 75, 1703–1711 (1994).
Kadonaga, J. T. & Kutach, A. K. The Downstream promoter element DPE appears to be as widely used as the TATA Box in Drosophila Core Promoters. Molecular and Cellular Biology. 20, 4754–4764 (2000).
Acknowledgements
The authors thank Indian council of Agricultural Research (ICAR) for providing funding to all the authors for carrying out this work.
Author information
Authors and Affiliations
Contributions
M.P. and B.R. conducted all experiments and wrote manuscript. B.S.Y. analyzed computational data. A.K. and M.S. helped in acquiring data. O.P.V. helped in editing manuscript. B.S. designed and supervised the study and also edited the manuscript. All authors reviewed and critically revised the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Pokhriyal, M., Ratta, B., Yadav, B.S. et al. Three newly identified Immediate Early Genes of Bovine herpesvirus 1 lack the characteristic Octamer binding motif- 1. Sci Rep 8, 11441 (2018). https://doi.org/10.1038/s41598-018-29490-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-29490-8
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.