Abstract
Patient-specific induced pluripotent stem cells (iPSCs) have the potential to be useful in the treatment of human diseases. While prior studies have reported multiple methods to generate iPSCs, DNA methylation continues to limit the totipotency and reprogramming efficiency of iPSCs. Here, we first show the competency of embryonic germ cells (EGCs) as a reprogramming catalyst capable of effectively promoting reprogramming induced by four defined factors, including Oct4, Sox2, Klf4 and c-Myc. Combining EGC extracts with these four factors resulted in formation of more embryonic stem cell-like colonies than did factors alone. Notably, expression of imprinted genes was higher with combined induction than with factors alone. Moreover, iPSCs derived from the combined inductors tended to have more global hypomethylation. Our research not only provides evidence that EGC extracts could activate DNA demethylation and reprogram imprinted genes, but also establishes a new way to enhance reprogramming of iPSCs, which remains a critical safety concern for potential use of iPSCs in regenerative medicine.
Similar content being viewed by others
Introduction
Induced pluripotent stem cells (iPSCs) represent a monumental scientific breakthrough in stem cell biology and regenerative medicine1,2, capable of breaking down various ethical and logistical obstacles associated with human embryonic stem cell (ESC) research3,4. iPSCs are generated by inducing the four “Yamanaka” transcription factors Oct4, Sox2, Klf4 and c-Myc (OSKM) into somatic cells5,6; and essentially, reprogramming is an epigenetic process for changing the fate of cells7,8,9. It involves a number of different mechanisms to overcome the epigenetic barriers that are imposed during differentiation10,11,12.
DNA methylation is a major handicap to reprogramming, causing both low efficiency of somatic cell reprogramming and instability of resulting pluripotent cells13,14. Previous studies have shown that differentiation-induced de novo DNA methylation can repress a large set of pluripotency genes including Oct4 and Nanog; whereas, active DNA demethylation is required for reactivation of pluripotency gene15,16,17. Furthermore, treatment of somatic cells with compounds that promote DNA demethylation facilitates the complete conversion of partially reprogrammed cells that would otherwise fail to reprogram into a pluripotent state11,14. Collectively, this research indicates that by interfering with repressive mechanisms, i.e. DNA methylation, the efficiency of transcription factor-induced reprogramming can be improved18,19.
Notably, DNA demethylation appears to be responsible for an increase in the pluripotency of extract-treated cells20,21,22. Reprogramming using extracts involves reversible permeabilization of somatic cells followed by exposure to extracts. Using this approach, several pluripotent cell types, including ESCs23,24,25,26 and embryonal carcinoma cells23,24,25,26,27, have been shown to elicit changes in the cell fate of somatic cells. Indications of reprogramming in this system include induction markers of pluripotency and downregulation of lamin A. More importantly, OCT4 activation is associated with DNA demethylation in the OCT4 promoter23; the NANOG promoter appears to be more readily demethylated, because Nanog overcomes reprogramming barriers and induces pluripotency in minimal conditions28. Observed alterations in the expression profiles of reprogrammed cells imply epigenetic modifications on DNA have taken place. Nevertheless, demethylation is incomplete and not all regions examined on OCT4 are equally demethylated29,30, in contrast to what is seen in ESCs or carcinoma cells. In the mouse embryos, migrating primordial germ cells (PGCs) reach the gonads at around 10.5 dpc. They undergo an extensive active genome-wide DNA demethylation, including erasure of genomic imprints. This rapid demethylation process is complete by 13.5 dpc31,32,33. Derived from PGCs, embryonic germ cells (EGCs) are pluripotent and harbor an epigenome similar to that of PGCs34,35. Studies have shown that EGC–thymocyte hybrids induce pluripotency markers and can differentiate into all three germ layers in chimera, which are characterized by demethylation of several non-imprinted and imprinted genes36. Furthermore, EGCs contain a substance with discrete roles in cell-fuse-mediated pluripotent reprogramming and imprint erasure in somatic cells37,38.
Genomic imprinting is an epigenetic alteration through which gene expression is regulated in a monoallelic manner. Abnormal expression of imprinted genes disrupts fetal development and is associated with both genetic diseases and malignancies39,40. Aberrant expression of imprinted genes has been observed with reprogramming of somatic cells by nuclear transfer41,42 or viral-mediated factors43,44,45. The methylation abnormalities in these cells result from the incomplete reprogramming. EGC fusion reportedly resets the epigenetic reprogramming of both imprinted and non-imprinted genes, which supports full reprogramming36. Yet, the precise mechanism affecting reprogramming remains unclear.
Based on the studies outlined above, we speculate that EGC extracts could enhance reprogramming by its unique capacity to actively drive the DNA demethylation process; however, the exact degree of reprogramming is unclear. Thus, we examined the reprogramming ability and mechanism of EGC extracts, which may have the potential to provide highly efficient and safe iPSCs.
Results
Treatment with EGC extracts enhanced generation of iPSCs
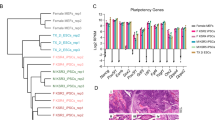
To investigate the role of EGC extracts in reprogramming, we first established EGCs from E12.5 embryos such that EGCs closely resembled ESCs with regard to pluripotency and related marker expression (Supporting Information Fig. S1). Next, we examined the effect of EGC extracts on MEFs infected with the four reprogramming factors. As described in Fig. 1A, the induction process involves infection of MEFs with retroviruses encoding the four factors with or without EGC extracts for 1 h and culture in ESC medium thereafter. As shown by FACS (Fig. 1C), percentage of S phase cells (36.7%, 78.11%) in the combined group (EGC extract plus OSKM treatment) was remarkably higher than that in the group without EGC extract treatment (19.68%, 30.48%) on 3 and 6 days after treatment. Proliferation activity [cell cycle proliferation index, PI (%)] of cells in the combined group was higher than that observed in the group without EGC extract treatment. Further, the morphology and growth of cells changed quickly (Fig. 1B). iPSC colonies were observed in the combined group first (at day 5) and the number of colonies increased with time in culture; whereas, iPSC colonies could only be readily identified in the four-factor group at day 7. Moreover, the number of AKP-positive colonies in the combined group was 5–7 times higher than in the four-factor treated group (Fig. 1D). As previously reported, an ESC-like morphology was used to identify putative iPSC colonies and the efficiency was roughly determined by number of AKP-positive colonies; thus, our results show that treatment with EGC extracts enhances four-factor mediated reprogramming.
Treatment with embryonic germ cell (EGC) extracts enhances the generation of induced pluripotent stem cells (iPSCs). (A) Schematic representation of iPSC generation from mouse embryonic fibroblasts (MEFs) by ectopic expression of Oct4, Sox2, Klf4 and c-Myc (OSKM) followed by exposure to EGC extracts. (B) Sequential morphology of iPSC generation with EGC extract treatments (a) of MEFs (b–d) Morphology of epithelium-like cells after infection at day 3 (e) embryonic stem cell-like colonies formed at day 5 (f) and as colonies become bigger at day 7. (C) Left: Cell cycle by FACS at day 3 and 6 from four factor-infected MEFs treated with EGC extracts and four factor-infected MEFs. Right: Percentage of cells in each cell cycle phase and PI (cell cycle proliferation index) in OSKM transfected MEFs with or without EGC extracts treatment. Mean values ± standard error of the mean (SEM) of three independent experiments are shown. (D) Left: Representative images of alkaline phosphatase-positive (AKP + ) colonies. Cells were fixed at day 8. Right: Number of AKP + colonies. Mean values ± SEM of a representative experiment are shown, n = 3. Scale bar = 100 µm.
iPSCs induced by both EGC extracts and four factors are pluripotent
iPSCs were generated in both the combined and four-factor groups, named EG-4F-iPSCs and 4F-iPSCs, respectively. EG-4F-iPSCs exhibited strong AKP activity (Fig. 2A-b) and expressed pluripotency markers, such as Oct4 and SSEA1 (Fig. 2E), in immunofluorescence analysis. Karyotype analysis exhibited normal 40 XX chromosomes (Fig. 2A-c). Real-time PCR analysis indicated that expression levels of pluripotency marker genes in EG-4F-iPSCs, including Oct4, Nanog, Sox2 and Rex1, were similar to those of 4F-iPSCs and markedly elevated compared with levels in MEFs (Fig. 2B).
Expression of pluripotent genes, In vitro and in vivo differentiation and the of iPSCs generated by EGC extract and OSKM combination. (A-a) Morphology of EG-4F-iPSCs P15. (A-b) Alkaline phosphatase-positive colonies and (A-c) normal karyotype (20pairs). (B) Quantitative PCR analysis of pluripotent markers Oct4, Nanog, Rex1 and Sox2 in EG-4F-iPSCs. Error bars indicate standard error of the mean (n = 3). Results were normalized to GAPDH expression. *P < 0.01, versus other cells. (C-a) Embryoid body formation by suspension culture. (C-b) Teratomas. (C-c) RT-PCR analysis of germ layer markers. The grouping blots of germ layer markers were cropped from different parts of the same gel. The grouping blots of gapdh were cropped from different parts of the same gel. (D) Hematoxylin and eosin staining of teratomas. Scale bar = 50 µm. (E) Immunofluorescence staining of pluripotent genes.
We also examined the differentiation capacity of EG-4F-iPSCs in vitro and in vivo (Fig. 2C,D). Similar to 4F-iPSCs, EG-4F-iPSCs form EBs in suspension culture and EBs expressed the three germ layer markers, Pax6 (ectoderm), eomes (mesoderm), and TTR (endoderm), as seen by RT-PCR (Figs 2C and S3). Further, EG-4F-iPSCs injected into nude mice formed teratomas after 6 weeks. Histological examination of resulting teratomas revealed tissues originating from all three embryonic germ layers. Taken together, these results indicate there is no significant difference in self-renewal or differentiation capacity between EG-4F-iPSCs and 4F-iPSCs.
EGC extracts promoted DNA demethylation in OSKM treated cells
It has been suggested that epigenetic reprogramming might be necessary for the embryonic genome to return to a pluripotent state. As such, we were interested in comparing global methylation patterns between EG-4F-iPSCs and 4F-iPSCs. Global MeDIP analysis allowed for identification of the baseline between methylated and unmethylated sequences.
We found changes in both hypo- and hypermethylation patterns took place throughout the genome between the two groups (Fig. 3A). The number of hypomethylated peaks was 32,783 in EG-4F-iPSCs, which was much higher than observed in 4F-iPSCs (8,951). This shows that EGC extracts could dramatically reduce methylation levels.
Genomic methylation patterns. (A) Hypo- and hypermethylation peak counts obtained from three cell lines in each group. EG-4F-iPSC versus 4F-iPSC peaks were considered as hypermethylation peaks; 4F-iPSC versus EG-4F-iPSC peaks were considered as hypomethylation peaks. (B) Average proportions of peaks within each region as defined by genomic structure. *P < 0.05 (C) Average proportions of peaks within each region as defined by distance from the CpG island. (D) Enrichment analysis of hypomethylated genes covered with peaks. (E) Comparison of imprinted gene expression in EG-4F-iPSCs and 4F-iPSCs as quantified by mRNA expression. Error bars indicate standard error of the mean (n = 3). Results were normalized to human histone H2A.Z (H2AFZ) expression. (F) Bisulfite sequencing of Nanog and OCT4 promoter region. Black circles represent methylated sites, white circles represent unmethylated sites. Global methylated cytosines are shown as %M.
We then scrutinized DNA hypo- and hypermethylation distribution in the context of gene structure. Four relevant regions were defined: promoter, gene body, transcriptional termination region (TTR) and the intergenic region. DNA hypo- and hypermethylation were distributed differently in the four regions. As shown in Fig. 3B, hypermethylation was concentrated mainly within gene body areas and could also be detected in the intergenic region, suggesting EGC extracts could selectively induce different methylation modifications. Next, we examined methylation changes in four regions defined by their distance from CpG islands: the CpG island itself, Shore, Shelf and Open Sea regions; the latter three regions were 2 kb, 2–4 kb, and more than 4 kb from the CpG island, respectively. Most hypo- and hypermethylation patterns were detected in the Open Sea region. No differences in the distribution of hypo- and hypermethylation status were found in any of these four regions (Fig. 3C).
In addition, we considered hypomethylated genes with peak enrichment analysis (Fig. 3D). Differentially hypomethylated genes were primarily involved in cellular processes, metabolism, and biological processes.
Furthermore, demethylation of OCT4 and Nanog promotors was detected by bisulfite sequencing. As shown in Fig. 3F, the Nanog promoter region was highly methylated in MEFs (74%) and almost completely unmethylated in B6ES (2%). A similar methylation pattern was observed in OCT4 examined region, where 89% of the CpG sites were methylated in MEFs but only 13% were methylated in B6ES. EGCs extraction treatment (EG-4F-iPS N8) was shown to decrease the methylation levels of both regions (6% and 18%, respectively) than that of OSKM induction without EGCs treatment (14% and 27% respectively in 4F-iPS N2).
Expression of imprinted genes increased after EGC extract treatment
Previous studies have demonstrated that methylation of imprinted genes could be erased during hybridization by fusion of thymic lymphocytes to EGCs33. To determine whether EGC extracts were also capable of eliciting epigenetic modifications on imprinted genes, we examined imprinted gene expression by real-time PCR. Figure 3E shows upregulation of both maternally expressed (Igf2r, Gtl2 and H19) and paternally expressed (Igf2, Snrpn) imprinted genes in EG-4F-iPSCs compared with 4F-iPSCs. Increasing expression of imprinted genes suggested exposure of MEF nuclei to EGC extracts could induced a more reprogrammable state.
EGC extracts can partially reprogram MEFs
Because EGC extracts could enhance reprogramming of iPSCs with epigenetic modifications, we investigated the direct reprogramming roles of extracts on MEFs. We exposed reversibly permeabilized MEFs to EGC or MEF extracts (control). Cells were plated and the number of cells was determined from day 1 to day 6 after treatment, as shown in Fig. 4A–E. Our results indicate EGC extracts increase the growth rate of MEFs relative to the control, with a peak at day4 (Fig. 4A). Q-PCR analysis suggested that expression levels of Oct4, Nanog and Sox2 with extract treatment increased at first and then decreased, reaching a peak at day 4 consistent with observed changes in growth rate (Fig. 4E). Moreover, EGC extracts can induce the formation of a few colonies resembling ESC morphology as early as day 5 post-treatment (Fig. 4B), whereas, MEFs treated with MEF extracts did not show any morphological changes (data not shown). These colonies displayed positive AKP (Fig. 4C) and endogenous expression of Oct4 (Fig. 4D), but the colonies could not be passaged.
Reprogramming of permeabilized MEFs induced by EGC extracts. (A) Proliferation rate of EGC extract-treated MEFs compared with MEF extract-treated MEFs at different days. Error bars represent standard error of the mean (SEM; n = 3). (B a–d) Morphology of colonies formed following treatment of MEFs with EGC extracts. (C) Induction of alkaline phosphatase activity in MEFs after exposure to EGC extracts. (D a-b) Immunofluorescence staining of Oct4 positive colonies following exposure of MEFs to EGC extracts. Scale bars = 25 µm. (E) Quantitative PCR analysis of pluripotency marker and laminA expression in MEFs after incubation with EGC extracts. Error bars indicate SEM (n = 3). Results were normalized to GAPDH expression. *P < 0.05, versus control. (F) Q-PCR analysis of imprinted gene expression between EGC extract-treated MEFs and MEF extract-treated MEFs at 4 days after treatment. Results were normalized to human histone H2A.Z (H2AFZ) expression. *P < 0.05, **P < 0.01, versus control.
We next examined the expression of imprinted genes between MEF and EGC extract-treated cells by qPCR analysis. As shown in Fig. 4F, maternally expressed imprinted genes Igf2r and H19 were increased in EGC extract-treated cells, most especially H19, which exhibited remarkably higher expression. Moreover, paternally expressed imprinted genes Igf2 and Snrpn were also upregulated. These results demonstrate MEFs can be programmed by EGC extracts. Unfortunately, it is difficult to realize full reprogramming with extracts alone.
Discussion
Our data indicate that EGC extracts improved reprogramming and facilitated the establishment of a pluripotent state in somatic cells. Experiments using somatic cell nuclear transfer46, cell fusion36,37,38 and extract-mediated22,23,26 induction confirmed that pluripotent cells, similar to oocytes, contain a lot of unknown factors capable of affecting somatic cell reprogramming. Previous reports have shown the roles of ESC, embryonal carcinoma and EGC extracts on reprogramming by hybrids with somatic cells; notably, the roles of EGCs in epigenetic reprogramming trumped the others36,47,48. Derived from PGCs, EGCs harbor an epigenome similar to that of PGCs, which show a genome-wide reprogramming of DNA methylation34,35; thus, EGCs might still contain demethylation factors and be better for reprogramming than ESCs. As such, we performed reprogramming using EGC extracts in the present study instead of ESC extracts.
As we expected, when OSKM induced MEFs were exposed to EGC extracts, we observed increased expression of pluripotent genes, accelerated appearance and number of ESC-like colonies, and significantly improved reprogramming efficiency. Previous achievement of reprogramming was restricted only to fusion of somatic cells and EGCs22. Here, we provide a new platform to improve reprogramming.
In addition, we found imprinted gene expression was also increased in EGC extract-treated cells. Genomic imprinting is essential for normal mammalian development and shows a substantial degree of stability in ESCs49. Further, abnormal allelic expression of imprinted genes in iPSCs has previously been shown to correlate with extensive methylation43,44, which could cause aberrant cell differentiation and obstruct transformation of iPSC lines to be used for regenerative medicine.
With further analysis of whole-genome methylation, we found that global demethylation was lower in EG-4F-iPSCs than in 4F-iPSCs. And EGC extraction treatment was shown to increase the methylation levels of both Nanog and OCT4 promoter regions. These were consistent with previous reports which showed that methylation of imprinted or non-imprinted genes was erased in EGC-fused cells23. Reversion of nuclei also occurred after fusion with ESCs; however, methylation patterns of imprinted genes were maintained23,47. This suggested EG-4F-iPSCs acquired improved pluripotency with less epigenetic memory.
Unfortunately, compared with EGC extract and OSKM combination, which efficiently induced fully competent iPSCs, application of EGC alone was ineffective at reprogramming of MEFs. Such a disparity may reflect the importance of varying reprogramming kinetics among different proteins. Apparently, amounts and effective time of proteins in the EGC extract may be inadequate and could not reach the reprogramming threshold.
The present study explores EGC extract as a unique reprogramming catalyst. Combining EGC extracts with OSKM resulted in improved efficiency and quality of generated iPSCs with less epigenetic memory as indicated by actively drive the DNA demethylation process. The reprogrammed cells tended to have more global hypomethylation. Use of EGC extracts in reprogramming may provides new approach and research strategies to yield more efficient and safer iPSCs. Further identification of relevant demethylation factors and reprogramming factors in the EGC extracts may provide us better understanding of the PGC demethylation mechanism and a significant advance in generating iPSCs in a safer manner.
Materials and Methods
EGC line
Animal handling followed the National Institutes of Health guidelines, and experimental procedures were approved by the Harbin Medical University Ethical Review Committee. Mouse fetuses from C57BL/6 J females mated with DBA/2 J males were collected at 12.5 days of pregnancy (E12.5). Female mice were sacrificed and fetuses were dissected away from their extraembryonic membranes. Genital ridges were dissociated from the mesonephros and dorsal walls of embryos. Genital ridge tissue was washed once with calcium-free Dulbecco’s phosphate-buffered saline (DPBS) and incubated in trypsin/EDTA solution for 5–10 min at 37 °C. Next, PGCs were dissociated by gently disrupting the genital ridge and the resulting cell suspension was centrifuged at 250 × g for 5 min. The cell pellet was resuspended in KnockOut™ Dulbecco’s Modified Eagle’s Media (DMEM; Gibco, USA) containing 20% KnockOut Serum Replacement (Gibco), 2 mM L-glutamine, 1% nonessential amino acids (NEAA), 0.1 mM 2-mercaptoethanol (Gibco), 20 ng/ml recombinant human basic fibroblast growth factor (Invitrogen, USA), 1000 U/ml recombinant leukemia inhibitory factor (LIF; Invitrogen), 40 ng/ml stem cell growth factor (Peprotech, USA), 1% penicillin and streptomycin (P/S; Gibco). After culture for 7−10 days, colonies were picked and seeded into new wells containing feeder cells for subculture. After 2–3 days, cells were passaged and cultured in LIF-containing medium.
Cell culture
MEFs were isolated from E13.5 B6D2F1 mouse embryos and cultured in DMEM with 10% fetal bovine serum (FBS). R1 ESCs were cultured in DMEM with 15% ESC-qualified FBS (Invitrogen), 0.1 mM NEAA, 0.1 mM 2-mercaptoethanol and 1000 U/ml LIF. Plat-E cells (Cyagen, USA) were maintained in DMEM containing 10% FBS, 1% P/S, 1 mg/ml Puromycin (Sigma, USA) and 100 mg/ml blasticidin S (Merck, Germany).
Cell extract preparation
EGCs were washed in DPBS and centrifuged at 1000 rpm for 5 min at 4 °C. Next, cells were suspended in cold DPBS containing 1 mM dithiothreitol, 0.1 mM phenylmethylsulfonyl fluoride and 0.1 mM protease inhibitor cocktail, and then incubated for 3 h on ice. After incubation, cells were repeatedly frozen and thawed in liquid nitrogen. Cell lysates were then centrifuged at 25,000 rpm for 15 min at 4 °C. The pH and osmotic pressure of the supernatant were then adjusted to physiological levels, before the protein concentration of each batch of supernatant was measured using an ultraviolet spectrophotometer (Eppendorf AG 22331, Hamburg, Germany). Finally, extracts were frozen in liquid nitrogen for future research.
Permeabilization
MEFs were washed in Ca2+- and Mg2+- free DPBS, permeabilization cold transport buffer (110 mM potassium acetate, 5 mM sodium acetate, 2 mM magnesium acetate, 1 mM EGTA, 2 mM dithiothreitol, protease-inhibitor cocktail and 20 mM HEPES) containing 1–15 µg/ml digitonin and then placed on ice for 2 min. The permeabilization reaction was halted by adding 900 µl of transport buffer and cells were collected by centrifugation at 700 × g for 10 min at 4 °C. To estimate permeabilization efficiency (Supporting Information Fig. S2), treated cells were incubated with 10 µg/ml propidiumiodide (PI, Sigma) for 10 min. Permeabilization was assessed by monitoring uptake of a 10,000-molecular weight dextran Oregon green 488 (50 µg/mL; Invitrogen) and PI. To reseal plasma membranes, the cell suspension was diluted with DMEM containing 2 mM CaCl2.
Induction of pluripotent stem cells by EGC extracts and OSKM
The retroviral pMX vector with OSKM cDNAs was transfected into Plat-E cells using Lipofectamine® LTX (Invitrogen). Supernatants were collected at 48 h and used to infect MEFs. The second round infection occurred at 72 h. The following day (day 0), MEFs were permeabilized and suspended in MEF or EGC extracts containing an ATP-regenerating system. After resealing plasma membranes, treated MEFs were reseeded on a 24-well plate and cultured in ESC medium (DMEM medium supplemented with15% ESC-qualified FBS, NEAA, P/S). On day 3, transfected MEFs were seeded on a feeder layer. On day 4, medium was replaced with KnockOut DMEM medium containing 15% KnockOut Serum Replacement, 0.1 mM NEAA, 0.1 mM β-mercaptoethanol and 1000U/ml LIF. iPSC colonies were picked, expanded and passaged using 0.25% trypsin/ethylenediaminetetraacetic acid (EDTA) every 2–3 days on feeder layers in ESC medium.
In vitro and in vivo differentiation
The in vitro differentiation potential of iPSCs was evaluated by embryoid body (EB) formation. Cells were dissociated with trypsin and then cultured in low-attachment plates with ESC medium without LIF for 6 days. The differentiation capacity was examined by RT-PCR check of germ layer markers (exoderm: pax6, mesoderm: eomes, endoderm: ttr). For in vivo differentiation, cells were injected subcutaneously into nude mice (Vital River Laboratory Animal Technology Co. Ltd, China). Tumors were collected after 2 months and processed for paraffin sectioning, followed by hematoxylin and eosin staining.
Alkaline phosphatase, immunostaining and karyotype analyses
Alkaline phosphatase (AKP) activity was detected with an AKP stain kit (Beyotime, China). For immunofluorescence analysis, cells were fixed in 4% paraformaldehyde 15 min and rinsed with 0.25% Triton X-100 in DPBS. After blocking, cells were incubated with Oct4 and SSEA1 antibodies (Santa Cruz Biotechnology, USA). Secondary antibodies were conjugated to fluorescein isothiocyanate (FITC; Santa Cruz Biotechnology) and nuclei were counter stained with Hoechst 33342 (Sigma).
For karyotype analysis, cells were incubated with colchicine for 1.5 h and then exposed to a hypotonic treatment with 0.56% KCl solution for 15 min. Fixation was performed with methanol and glacial acetic acid mixture (3:1). Finally, cell spreads were stained with Giemsa (Sigma) and separate metaphase karyotypes were counted.
Real-time PCR and RT-PCR
Total RNA was extracted from cells with TRIzol® reagent (Invitrogen) according to the manufacturer’s protocol. cDNA synthesis was achieved using oligo (dT) and SuperScript® reverse transcriptase (Takara, Dalian, China). Real-time PCR was performed using TransStart™ Top Green qPCR Super Mix Kit (TransGen, China). Relative amount of gene expression was analyzed by the 2-dd Ct method50. RT-PCR was performed using High Capacity cDNA Reverse Transcription Kit (ABI, American). Primer sequences are listed in Supplemental Method Table 1.
Whole-genome methylation analysis
DNA (3 μg) was sonicated at intensity 4 for 200 cycles per burst for 55 s in a Covaris S2. DNA fragments were end-repaired, ATP-tailed and adapter ligated with a Sample Preparation Kit (Illumina, USA). Next, DNA was recovered by AMPure® XP Beads and prepared for methylated DNA immunoprecipitation (MeDIP) using a Magnetic Methylated DNA Immunoprecipitation Kit (Diagenode, Belgium) according to the manufacturer’s protocol. After MeDIP, remaining DNA was PCR-amplified with sequencing primers and used for sequencing (Illumina HiSeq. 2500, USA). Raw reads were preprocessed using the FASTX-Toolkit. MeDIP-seq peaks were counted using MACS. EG-4F-iPSC versus 4F-iPSC (control) peaks were considered as hypermethylation peaks; whereas, EG-4F-iPSC versus 4F-iPSC (control) negative peaks were considered as hypomethylation peaks. Genes covered by peak-enrichment analysis were assessed using GO stats software (Bioconductor).
Bisulfite treatment and bisulfite sequencing
Demethylation of OCT4 and Nanog promotors was detected by bisulfite sequencing, in accordance with the method of Christel T. et al.26 Converted DNA were amplified by PCR using the primers listed below. The annealing temperatures were 60 °C. The PCR products were the cloned into the pGEM-T Easy Vector (Prpmega) system for sequencing. Primer pairs used were: OCT4 forward: 5′-GAGGTGCAATGGCTGTCTTGT-3′, reverse: 5′-ACCAACCAGTTGCTCGGATGC-3′; Nanog forward: 5′-GGTAGGGTAGGAGGTTTGAGG-3′, reverse: 5′-AAGGTTTTAGGCAACAATTAA-3′.
Statistical analysis
All data were obtained from at least three independent experiments. Statistical analysis of the data was performed with a Student’s t-test. P < 0.05 was considered statistically significant. All data are shown as mean ± standard deviation.
References
Yamanaka, S. The winding road to pluripotency (nobel lecture). Angewandte Chemie - International Edition 52, 13900–13909 (2013).
Okita, K. & Yamanaka, S. Induced pluripotent stem cells: opportunities and challenges. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 366, 2198–207 (2011).
de Miguel-Beriain, I. The ethics of stem cells revisited. Advanced Drug Delivery Reviews 82, 176–180 (2015).
Xu, X. et al. Prevention of β-amyloid induced toxicity in human iPS cell-derived neurons by inhibition of Cyclin-dependent kinases and associated cell cycle events. Stem Cell Res. 10(2), 213–227 (2013).
Takahashi, K. & Yamanaka, S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 126, 663–676 (2006).
Takahashi, K. et al. Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell 131, 861–872 (2007).
Watanabe, A., Yamada, Y. & Yamanaka, S. Epigenetic regulation in pluripotent stem cells: a key to breaking the epigenetic barrier [review]. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 368, 20120292 (2013).
Maherali, N. et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell 1, 55–70 (2007).
Papp, B. & Plath, K. Reprogramming to pluripotency: stepwise resetting of the epigenetic landscape. Cell Res 21, 486–501 (2011).
Xie, B. et al. Histone H3 lysine 27 trimethylation acts as an epigenetic barrier 1 in porcine nuclear reprogramming. Reproduction 151, 9–16 (2015).
Han, J., Sachdev, P. S. & Sidhu, K. S. A combined epigenetic and non-genetic approach for reprogramming human somatic cells. PLoS One 5, e12297 (2010).
Djuric, U. & Ellis, J. Epigenetics of induced pluripotency, the seven-headed dragon. Stem Cell Res. Ther. 1, 3 (2010).
De Carvalho, D. D., You, J. S. & Jones, P. A. DNA methylation and cellular reprogramming. Trends in Cell Biology 20, 609–617 (2010).
Bhutani, N. et al. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature 463, 1042–7 (2010).
Kellner, S. & Kikyo, N. Transcriptional regulation of the Oct4 gene, a master gene for pluripotency. Histol. Histopathol. 25, 405–412 (2010).
Yang, J., Gao, C., Chai, L. & Ma, Y. A novel SALL4/OCT4 transcriptional feedback network for pluripotency of embryonic stem cells. PLoS One 5, e10766 (2010).
Fernández-Arroyo, S. et al. Activation of the methylation cycle in cells reprogrammed into a stem cell-like state. Oncoscience 2, 958–967 (2016).
Wang, Q. et al. Lithium, an anti-psychotic drug, greatly enhances the generation of induced pluripotent stem cells. Cell Res 21, 1424–1435 (2011).
Stadtfeld, M. et al. Ascorbic acid prevents loss of Dlk1-Dio3 imprinting and facilitates generation of all-iPS cell mice from terminally differentiated B cells. Nat. Genet. 44, 398–405, S1-2 (2012).
Han, J. & Sidhu, K. Embryonic stem cell extracts: use in differentiation and reprogramming. Regen. Med. 6, 215–27 (2011).
Bui, H.-T. et al. Epigenetic reprogramming in somatic cells induced by extract from germinal vesicle stage pig oocytes. Development 139, 4330–40 (2012).
Xiong, X. R. et al. Cellular extract facilitates nuclear reprogramming by altering DNA methylation and pluripotency gene expression. Cell Reprogr. 16, 215–222 (2014).
Taranger, C. K. et al. Induction of dedifferentiation, genomewide transcriptional programming, and epigenetic reprogramming by extracts of carcinoma and embryonic stem cells. Mol Biol Cell 16, 5719–5735 (2005).
Xu, Y. N. et al. ES cell extract-induced expression of pluripotent factors in somatic cells. Anat. Rec. 292, 1229–2234 (2009).
Kwon, Y. W. et al. Role of Zscan4 in secondary murine iPSC derivation mediated by protein extracts of ESC or iPSC. Biomaterials 59, 102–115 (2015).
Cho, H. J. et al. Induction of pluripotent stem cells from adult somatic cells by protein-based reprogramming without genetic manipulation. Blood 116, 386–395 (2010).
Freberg, C. T., Dahl, J. A., Timoskainen, S. & Collas, P. Epigenetic reprogramming of OCT4 and NANOG regulatory regions by embryonal carcinoma cell extract. Mol. Biol. Cell 18, 1543–53 (2007).
Theunissen, T. W. et al. Nanog overcomes reprogramming barriers and induces pluripotency in minimal conditions. Curr. Biol. 21, 65–71 (2011).
Nishino, K. et al. DNA methylation dynamics in human induced pluripotent stem cells over time. PLoS Genet. 7, (2011).
Ohi, Y. et al. Incomplete DNA methylation underlies a transcriptional memory of somatic cells in human iPS cells. Nat Cell Biol 13, 541–549 (2011).
Hajkova, P. et al. Epigenetic reprogramming in mouse primordial germ cells. Mech Dev. 117, 15–23 (2002).
Tang, W. W. C. et al. A unique gene regulatory network resets the human germline epigenome for development. Cell 161, 1453–1467 (2015).
Messerschmidt, D. M., Knowles, B. B. & Solter, D. DNA methylation dynamics during epigenetic reprogramming in the germline and preimplantation embryos. Genes and Development 28, 812–828 (2014).
Nagamatsu, G. et al. Induction of pluripotent stem cells from primordial germ cells by single reprogramming factors. Stem Cells 31, 479–87 (2013).
Bazley, F. A. et al. Direct Reprogramming of Human Primordial Germ Cells into Induced Pluripotent Stem Cells: Efficient Generation of Genetically Engineered Germ Cells. Stem Cells Dev. 24, 2634–48 (2015).
Tada, S., Tada, T., Lefebvre, L., Barton, S. C. & Surani, M. A. Embryonic germ cells induce epigenetic reprogramming of somatic nucleus in hybrid cells. EMBO J. 16, 6510–6520 (1997).
Ficz, G. & Reik, W. Reprogramming by cell fusion: Boosted by tets. Molecular Cell 49, 1017–1018 (2013).
Piccolo, F. M. et al. Different Roles for Tet1 and Tet2 Proteins in Reprogramming-Mediated Erasure of Imprints Induced by EGC Fusion. Mol. Cell 49, 1023–1033 (2013).
Horsthemke, B. & Wagstaff, J. Mechanisms of imprinting of the Prader-Willi/Angelman region. American Journal of Medical Genetics, Part A 146, 2041–2052 (2008).
Quinn, E. M. et al. Transcriptome Analysis of CD4+T Cells in Coeliac Disease Reveals Imprint of BACH2 and IFNγ Regulation. PLoS One 10, e0140049 (2015).
Xu, W. et al. Effects of DNMT1 and HDAC Inhibitors on Gene-Specific Methylation Reprogramming during Porcine Somatic Cell Nuclear Transfer. PLoS One 8, (2013).
Stadtfeld, M. et al. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature 465, 175–81 (2010).
Liu, L. et al. Activation of the imprinted Dlk1-Dio3 region correlates with pluripotency levels of mouse stem cells. J. Biol. Chem. 285, 19483–90 (2010).
Zhou, W. et al. Higher methylation in genomic DNA indicates incomplete reprogramming in induced pluripotent stem cells. Cell. Reprogram. 15, 92–9 (2013).
Chang, G. et al. High-throughput sequencing reveals the disruption of methylation of imprinted gene in induced pluripotent stem cells. Cell Res. 24, 293–306 (2014).
No, J. G. et al. Cell-free extract from porcine induced pluripotent stem cells can affect porcine somatic cell nuclear reprogramming. J. Reprod. Dev. 61, 90–98 (2015).
Battulin, N. R. et al. Reprogramming somatic cells by fusion with embryonic stem cells does not cause silencing of the Dlk1-Dio3 region in mice. World J. Stem Cells 4, 87–93 (2012).
Serov, O. L., Matveeva, N. M. & Khabarova, A. A. Reprogramming Mediated by Cell Fusion Technology. Int. Rev. Cell Mol. Biol. 291, 155–190 (2011).
Takikawa, S. et al. Human and mouse ZFP57 proteins are functionally interchangeable in maintaining genomic imprinting at multiple imprinted regions in mouse ES cells. Epigenetics 8, 1268–1279 (2013).
Chang, S., Chen, W. & Yang, J. Another formula for calculating the gene change rate in real-time RT-PCR. Mol. Biol. Rep. 36, 2165–2168 (2009).
Acknowledgements
This research was supported financially by National Science Foundation of China (31671545, 81772498), the State Key Development Program of Basic Research of China (2012CBA01303), Heilongjiang Province Science Funds for Distinguished Young Scientists (JC2015019), The Innovative Fund of Harbin Medical University Graduate Student (YJSCX2011-321HLJ).
Author information
Authors and Affiliations
Contributions
J.H., Q.S.Z., N.L., Y.L.G., R.Z.S. and Y.S.W. performed experiments. J.H., Y.K.F. and L.D. performed bioinformatics analysis. J.H., Z.Y.S. and L.L. designed experiments and wrote the manuscript. All authors approved its final version.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hu, J., Zhao, Q., Feng, Y. et al. Embryonic germ cell extracts erase imprinted genes and improve the efficiency of induced pluripotent stem cells. Sci Rep 8, 10955 (2018). https://doi.org/10.1038/s41598-018-29339-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-29339-0
This article is cited by
-
The extracts of osteoblast developed from adipose-derived stem cell and its role in osteogenesis
Journal of Orthopaedic Surgery and Research (2024)
-
Exploring the promising potential of induced pluripotent stem cells in cancer research and therapy
Molecular Cancer (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.