Abstract
Recent studies have revealed the importance of rare variants in myocardial infarction (MI) susceptibility in European populations. Because genetic architectures vary in different populations, we investigated how they contribute to MI susceptibility in Japanese subjects. We performed targeted sequencing of 36 coronary artery disease risk genes, identified by genome-wide association studies, in 9,956 cases and 8,373 controls. Gene-based association tests identified significant enrichment of rare variants in LDLR and PCSK9 in MI cases. We identified 52 (novel 22) LDLR variants predicted to be damaging. Carriers of these variants showed a higher risk of MI (carriers/non-carriers 89/9867 in cases, 17/8356 controls, OR = 4.4, P = 7.2 × 10−10), higher LDL-cholesterol levels and younger age of onset for MI. With respect to PCSK9, E32K carriers showed higher LDL-cholesterol levels and younger age of onset for MI, whereas R93C carriers had lower LDL-cholesterol levels. A significant correlation between LDL-cholesterol levels and onset age of MI was observed in these variant carriers. In good agreement with previous studies in patients with familial hypercholesterolaemia, our study in the Japanese general population showed that rare variants in LDLR and PCSK9 were associated with the onset age of MI by altering LDL-cholesterol levels.
Similar content being viewed by others
Introduction
Despite advances in therapeutic strategies, myocardial infarction (MI) remains a leading cause of morbidity and mortality worldwide1. To clarify the complex heritability of MI2, large-scale genome-wide association studies (GWAS) were performed to identify more than 160 susceptibility loci for coronary artery disease (CAD)3,4,5,6,7,8. Moreover, resequencing analyses9,10,11,12,13 have revealed that rare variants in lipid-related genes contribute to the susceptibility for MI.
Although genes associated with diseases are shared among populations, disease-associated rare variants are subject to variation depending on the population. For example, a genomic analysis using an exome array demonstrated that the association of low-frequency variants with blood lipids or CAD was different between participants of European ancestry and African ancestry13. The protective effect of ANGPTL4 low-frequency variant on CAD was reported in a European population10,11 but was not observed in a Chinese population12. Thus, these uneven distributions of rare variants could be explained by population differences. Therefore, their effects should be evaluated using a large number of samples in each population to successfully develop population-specific precision medicine, in which the most appropriate preventive therapy could be chosen based on a population-specific genetic risk profile.
Additionally, judging from the recent findings10,11,12,13 that significant rare variants associated with CAD reside in GWAS-identified genes for CAD (e.g., LDLR, PCSK9, APOB), the GWAS-identified genes could be good targets for rare variant discovery. The significance of GWAS-identified genes in rare and functional variant discovery has also been demonstrated in other studies on dyslipidaemia14,15. Based on these genetic findings, to detect efficiently the rare variants associated with CAD, we adopted a strategy for performing targeted sequencing of 36 genes from CAD-associated GWAS loci reported up to the beginning of our present study and conducted an association analysis using 9,956 cases and 8,373 controls in the Japanese population. The aim was to better understand the contribution of rare variants to the susceptibility of MI, followed by proposing a possible preventive strategy for Japanese.
Results
Summary Of Two-Stage Targeted Sequencing
In the discovery stage, targeted sequencing of 36 genes (90,823 bp) was performed in 2,811 cases and 2,974 controls (Table 1) and covered 98.9% of targeted bases with a minimum of 20-fold depth (DP) (Supplementary Fig. S1). After QC, 1,630 variants (minor allele frequency (MAF) < 0.05) were detected in 2,775 cases and 2,965 controls (Fig. 1) (Supplementary Tables S2 and S3). Of these variants, 1,235 were novel, among which 508 and 465 novel ones were observed only in cases and controls, respectively, and 262 novel ones were identified in both groups. After excluding the synonymous variants, we performed single-variant and gene-based association analyses using 1,021 single nucleotide variant (SNV) of missense and nonsense, indel frameshift and splice-site variants with a minor allele frequency (MAF) < 0.05. Single-variant association analysis identified 16 SNVs that showed P < 0.05 in Fisher’s exact test (Supplementary Table S4). Gene-based association was analysed with the Cohort Allelic Sum Test (CAST)16 and Sequence Kernel Association Test (SKAT)17, and we found 7 genes (PCSK9, GUCY1B3, PLG, ICA1L, NBEAL1, TCTN1 and LDLR) that showed P < 0.05 in at least one of the following three variant categories: (1) all non-synonymous variants; (2) damaging, defined by all disruptive (null) variants and missense variants annotated as deleterious by all five protein function prediction algorithms, PolyPhen-2 HumDiv, Polyphen2-HumVar18, SIFT19, MutationTaster20 and LRT21 score; and (3) disruptive (null) variants (nonsense, indel frameshift and splice-site variants) (Supplementary Table S5). In the replication stage, the 11 genes including the 16 SNVs and 7 genes that showed an association in the discovery stage were sequenced (39,944 bp) in 7,316 independent cases and 5,828 independent controls (Table 1), 98.2% of the targeted bases were covered with DP ≥ 20 (Supplementary Fig. S2), and 1,028 variants were identified in 7,181 cases and 5,408 controls after quality control (Fig. 1) (Supplementary Table S6). Of these variants, 791 were novel, among which 356 and 236 novel ones were observed only in cases and controls, respectively, and 199 novel ones were identified in both groups. Among 7 genes targeted in both the discovery and replication stages, 587 novel variants were identified in the discovery stage, and 118 were repeatedly identified in the replication stage. That is, 469 other novel variants were identified only in the discovery stage.
Overall design for the two-stage targeted sequencing study. Missense, nonsense, indel frameshift and splice-site variants with minor allele frequency less than 5% were tested after excluding the synonymous variants. In the single variant test, we set the study-wide significance threshold to P = 3.1 × 10−5. In the gene-based test, we set the study-wide significance threshold to P = 4.6 × 10−4.
Results of association test
In the meta-analysis of single-variant association tests (Table 2), we found one missense variant that showed a study-wide significant association with MI: chr1:g.55505604 G > A (PCSK9: E32K, OR = 1.7, P = 3.5 × 10−7).
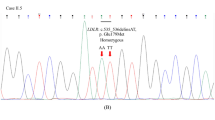
In a meta-analysis of gene-based association tests (Table 2), we identified significant associations in LDLR and PCSK9. In LDLR, we found a total of 138 non-synonymous variants, in which 52 damaging variants with 14 disruptive ones were identified. In damaging variants, 22 out of 52 were novel variants, while 6 out of 14 were novel in disruptive variants. All LDLR disruptive variants were confirmed by Sanger sequencing. The results of gene-based tests in the three variant categories are shown in Supplementary Table S7. The damaging variants showed significant enrichment (OR = 4.4, P = 7.2 × 10−10) in MI cases. With respect to the disruptive variants, we found a stronger genetic effect (OR = 15) on MI risk, but the association was weakened (P = 5.8 × 10−7), potentially due to the small number of samples (36 in cases and 2 in controls).
In PCSK9, we found 94 non-synonymous, 18 damaging and 6 disruptive variants. We found a significant association with MI in all non-synonymous PCSK9 variants by SKAT (P = 2.3 × 10−7), but that association was not detected by CAST. The damaging and disruptive variants did not show significant associations. Because we observed that E32K was a risk variant for MI whereas R93C was protective in the single-variant test (Supplementary Table S4), PCSK9 appears to harbour rare variants with opposite effects: deleterious and protective. To clarify the possibility that the co-existence of these opposite-effect variants might offset the association of PCSK9 variants with MI, we classified variants into a gain-of-function (GoF) group and a loss-of-function (LoF) group based on the Leiden Open (source) Variation Database(LOVD)22,23,24. As a result, we found significant associations in both the GoF group (OR = 1.3, P = 1.0 × 10−4) and the LoF group (OR = 0.7, P = 1.1 × 10−4). However, these associations were not significant after excluding E32K or R93C (P = 0.95 after excluding E32K in the GoF group and P = 0.70 after excluding R93C in the LoF group). These results suggest that E32K and R93C in PCSK9 have a predominant effect in the gene-based test for PCSK9. All tested categories of PCSK9 variants are shown in Supplementary Table S7.
In 5 MI cases, a damaging LDLR variant and PCSK9 E32K variant were detected, while this coexistence did not occur in any control subject.
Clinical phenotypes of rare variant carriers of LDLR and PCSK9
To assess the clinical impact of rare variants in LDLR and PCSK9, we examined the effects of these variants on serum LDL cholesterol levels and the onset age of MI. We also examined HDL cholesterol levels and triglycerides levels; however, we did not observe any statistically significant relationship with those variants.
When we compared LDL cholesterol levels among the three variant categories of LDLR in all subjects (cases and controls) whose prescription data and lipid profile data were both available (Table 3), LDL cholesterol levels in disruptive variant carriers were significantly higher than those in non-carriers, who did not have any LDLR or PCSK9 rare variants (+48.5 mg/dl, P = 7.6 × 10−13). LDL cholesterol levels were also higher in non-synonymous variant carriers and damaging variant carriers than in non-carriers (+5.0 mg/dl, P = 0.012, + 44.4 mg/dl, P = 6.9 × 10−17, respectively). Next, we examined the effect of LDLR rare variants on the age of MI onset in cases (Table 4). The onset age in disruptive variant carriers was significantly younger than those in non-carrier MI patients (−11.2 years, P = 5.2 × 10−10). The onset age in damaging carriers was also younger than that in non-carriers (−4.9 years, P = 1.3 × 10−3). With respect to PCSK9, we compared LDL cholesterol levels and the onset age of MI among the following groups: E32K carriers, R93C carriers and disruptive variant carriers. When we compared LDL cholesterol levels among these groups (Table 3), the E32K carriers showed higher LDL cholesterol levels than the non-carriers, who did not have any LDLR or PCSK9 variants (+18.0 mg/dl, P = 1.6 × 10−14), whereas R93C carriers had lower LDL cholesterol levels than non-carriers (−12.4 mg/dl, P = 5.6 × 10−5). In the disruptive variant group, LDL cholesterol levels were significantly lower than those in non-carriers (−38.5 mg/dl, P = 0.012). In the analysis of the onset age of MI in cases (Table 4), R93C carriers did not show a significant alteration (+0.22 years, P = 0.81). However, E32K carriers showed earlier onset than did non-carrier MI patients (−2.3 years, P = 3.7 × 10−4), whereas disruptive variant carriers showed later onset of MI (+8.2 years, P = 0.043). These results suggested that rare variants in LDLR and PCSK9 had a predominant effect on the onset age of MI in the MI patients. Three subjects who carried both E32K and R93C in PCSK9 were excluded from the analyses.
Both LDLR and PCSK9 are known to be causative genes for familial hypercholesterolemia (FH), which is a well-known risk for MI. The FH database23,24 contains some of the variants detected in this study in LDLR and PCSK9 (59/138 = 43% and 23/94 = 24%, respectively) (Supplementary Tables S8 and S9). To examine the effect of newly identified variants in this study, we removed these previously known variants from our data and explored how these associations changed among the three categories that showed a significant association with MI. As a result, each category showed drastic attenuation of signals (LDLR damaging P = 0.027, LDLR disruptive P = 0.13 and PCSK9 all non-synonymous (SKAT) P = 0.58), where statistical power was hampered, potentially due to the decreased number of variant carriers. After subtraction of these variant carriers, we had a limited number of samples (disruptive variant carriers in LDLR decreased from 38 to 6, damaging variant carriers in LDLR decreased from 106 to 24, and PCSK9 all non-synonymous variant carriers decreased from 4,748 to 277) (Supplementary Table S10). When we examined the LDL cholesterol levels and the onset age of MI using newly identified variants in LDLR and PCSK9 alone (Supplementary Tables S11 and S16), the effects of newly identified variants exhibited the same trend as those of previously known FH variants, where the statistical power was weakened, potentially due to the decreased number of variant carriers.
Dividing the study population according to lipid-lowering therapy or gender, subgroup analyses on the genetic effects of variants on serum LDL cholesterol levels or the onset age of MI were performed. As described above, the LDL cholesterol level analyses were conducted for all (case and control) subjects whose prescription data and lipid profile data were available, and the onset-age analyses were performed for all cases. In our study, the percentage of subjects treated by cholesterol-lowering medications was much greater in MI cases (47%) than in controls (9.1%), consistent with the standard LDL cholesterol-lowering therapy. Although the statistical power was weakened potentially due to the decreased number of carriers in each variant category, nearly the same trend was observed for the genetic effects of variants on serum LDL cholesterol levels in all subgroups (Supplementary Tables S12–15). However, for the genetic effects of variants on the onset age of MI, nearly the same trend was observed in subjects with cholesterol-lowering drugs (Supplementary Table S18) and male patients (Supplementary Table S19), whereas the trend was not observed in subjects without cholesterol-lowering drugs (Supplementary Table S17) and female patients (Supplementary Table S20).
To clarify the relationship between LDL cholesterol levels and MI onset in the association of LDLR and PCSK9 rare variants, we compared the changes in LDL cholesterol levels and changes in the onset age of MI estimated in the evaluation of the clinical phenotypes (Fig. 2). We found a significant linear correlation between these two factors in all variant categories (r = −0.94, P = 0.004). Moreover, a similar trend was observed in patients without previously known FH variants (r = −0.95, P = 0.054) (Supplementary Fig. S3). Dividing the study population according to lipid-lowering therapy or gender, subgroup analyses of this relationship were performed. A significant linear correlation was demonstrated in patients without cholesterol-lowering drugs (r = −0.96, P = 0.003), and a similar trend was observed in patients with cholesterol-lowering drugs (r = −0.85, P = 0.068) (Supplementary Fig. S4). In male patients, a significant linear correlation in all variant categories was observed (r = −0.91, P = 0.013), and a similar trend was observed in female patients (r = −0.79, P = 0.062) (Supplementary Fig. S5). Collectively, rare variants in these genes exert their effect on the onset age of MI in large part by altering LDL cholesterol levels.
Effects of LDLR and PCSK9 rare variants on LDL-C levels and onset age of MI. Dots represent the change from non-carriers of LDLR/PCSK9 rare variants for each group and lines indicate the 95% confidence interval. Effects were estimated using multiple linear regression models adjusted for age, gender, BMI, smoking status and cholesterol lowering medications in assessing LDL cholesterol levels, and controlling for the same parameters except age in assessing onset age of MI. Abbreviations: r: Pearson’s correlation coefficient.
Discussion
This study is the first to address the relationship between rare variants and MI in a large Japanese population. We identified a significant association of rare variants in LDLR and PCSK9 with MI. Our analysis revealed that rare variants in LDLR and PCKS9 affect onset age of MI by altering serum LDL cholesterol levels.
These dyslipidaemia-related genes are found to be susceptibility genes for MI in Japanese, which is consistent with observations made for different ethnic populations25,26,27. However, the contents of rare variants in these genes are different. When we examined the individual variants identified in the present study, 55 LDLR and 43 PCSK9 variants were not in the ExAC or FH databases, implying that they are novel and unique in the Japanese population (Supplementary Tables S8 and S9). In the Japanese population, judging from CAST and SKAT results, we observed a unidirectional effect of LDLR variants composed of loss-of-function variants with deleterious consequences for MI and a bidirectional effect of PCSK9 variants: PCSK9 E32K, which is well known as a FH-causing variant, is a gain-of-function variant with a deleterious effect for MI, while loss-of-function PCSK9 disruptive variants have a protective effect on the onset of MI. Indeed, the bidirectional effect of PCSK9 variants on both LDL cholesterol levels and MI risk were previously reported in studies of Mendelian dyslipidaemia28,29. Additionally, this bidirectional effect of PCSK9 variants on LDL cholesterol levels was observed in the general population30, but the relationship between the bidirectional effect and MI was not discussed. Hence, we are the first to present the bidirectional effect of PCSK9 rare variants on MI risk via LDL cholesterol levels in a population-based study.
We demonstrated a linear correlation between the changes in LDL cholesterol levels and changes in onset age of MI in carriers of LDLR or PCSK9 rare variants, which implies that rare variants in LDLR and PCSK9 influence the onset age of MI potentially by altering serum LDL cholesterol levels. This finding supports the idea that normalizing LDL cholesterol levels in carriers of LDLR or PCSK9 rare variants should be effective in preventing MI onset. A long-term cohort study provided supportive evidence indicating that statin therapy for normalizing LDL cholesterol levels in FH patients lowered the risk of MI onset to the same level as that in non-FH patients31. Therefore, given that carriers of LDLR or PCSK9 rare variants continue to be exposed to genetic effects after birth and accumulate the risk of MI, we propose that we should check LDLR and PCSK9 rare variants in patients with juvenile-onset hyper-LDL cholesterolaemia, whether a diagnosis of FH is made, and that a preemptive therapy for normalizing LDL cholesterol levels should be undertaken to prevent MI as long as patients have LDLR rare variants or PCSK9 gain-of-function variants. Notably, judging from our findings that the genetic effects of rare variants on LDL cholesterol level and onset age of MI were observed even in patients with cholesterol-lowering therapy, the ongoing lipid-lowering therapy in clinical practice might be insufficient to cancel the rare variants-associated hyper-LDL cholesterolaemia and MI.
Even with our careful curation of candidate genes for targeted sequencing, potential limitations remain. First, a study in a European population showed that rare alleles in APOA5 contributed to the risk for early onset MI25. However, we did not employ APOA5 because it did not meet our prespecified criteria. Second, although our gene selection was mainly based on the loci provided by the CARDIoGRAMplusC4D Consortium7. We performed the gene selection in 2013, after which the latest studies8,32 expanded the spectrum of GWAS loci for CAD and increased candidate genes for MI susceptibility. An additional analysis of rare variants in newly identified genes might be needed. Third, the classification into damaging variants and disruptive (null) ones was decided by prediction algorithms and not verified by experimental data. A similar classification was used in a previous study25 in which “deleterious (strict)” variants corresponded to our “damaging” variants. Fourth, we did not check and exclude subjects with FH, although our list for targeted sequencing included previously known FH genes. In our study, 43% of LDLR and 24% of PCSK9 rare variants were previously reported FH mutations. However, it is natural to find subjects with known FH mutations in the general population because other population-based studies have also identified variants previously described as causing FH25,27.
Despite these limitations, our analyses shed light on the Japanese-specific genetic architecture of MI risk driven by rare and low-frequency variants and elucidate a correlative link between LDL cholesterol level and onset age of MI in the presence of LDLR or PCSK9 rare variants. Recent parent-child genetic screening33 for previously documented 48 FH mutations (including 46 LDLR mutations, 1 APOB one, and 1 PCSK9 one) revealed a relatively low prevalence (0.8%) of such carriers. However, our targeted sequencing demonstrated that carriers of rare variants in dyslipidaemia-related genes are more prevalent in the general population. Compared with panel screening of previously reported FH mutations, wide screening using targeted sequencing might be more valuable because it could identify rare variants associated with LDL cholesterol levels and MI risk even if each variant has milder effects than those of known FH mutations. Given that dyslipidaemia-related genes could contribute to the pathogenesis of MI even under lipid-lowering therapy, more potent treatment of dyslipidaemia than ever should be recommended as a promising tool for prevention of MI. Identification of rare and low-frequency variants will provide a clue for clarifying the complex genetic architecture of MI risk as well as a rationale for the appropriate treatment against MI, which emphasizes the usefulness of targeted sequencing of candidate genes.
Methods
Study design
All methods were performed in accordance with the relevant guidelines and regulations.
A targeted sequencing was performed in two stages as shown in Fig. 1. In the discovery stage, we performed targeted sequencing in coding regions of 36 genes using 2,775 MI cases and 2,965 controls. We performed single-variant and gene-based association analysis and selected variants or genes that showed a P value of less than 0.05 in the discovery stage. These variants or genes were examined using 7,181 independent cases and 5,408 independent controls in the replication stage. After the two stages of sequencing, a meta-analysis was performed. We only used variants annotated as missense, nonsense, indel frameshift or splice-site with a minor allele frequency (MAF) of less than 5% throughout this study.
Study samples
All MI cases including discovery and replication stages were obtained from the BioBank Japan project34,35, which constructed a patient-oriented biobank that collected DNA samples from 200,000 patients suffering from at least one of 47 target diseases including MI between 2003 and 2008. As previously described36, cases in both stages were selected based on medical records and confirmed to satisfy both of the following criteria: (1) left ventricular wall motion abnormalities on echocardiography and (2) one or more coronary artery occlusion on angiography. If patients had experienced multiple MI events, the first episode was considered “onset of MI” in the onset-age analysis.
Controls in the discovery stage were collected from three different sites: Pharma SNP Consortium (PSC), Osaka-Midousuji Rotary Club (MRC) and the University of Tokyo Hospital. MRC and PSC samples were self-reported healthy volunteers. Controls from the University of Tokyo Hospital were examinees who underwent a health check-up, and individuals with a history of CAD were excluded. Controls for the replication stage were a mixture of cases registered in the Biobank Japan that had been used as GWAS controls in previous reports36,37. These control subjects consisted of patients with 5 diseases (cerebral aneurysm, oesophageal cancer, endometrial cancer, chronic obstructive pulmonary disease and glaucoma). Individuals with CAD were excluded from controls. All individuals were of Japanese ancestry and provided written informed consent to participate in this study. This study was approved by the ethics committees of the University of Tokyo and RIKEN Center for Integrative Medical Sciences.
Gene selection
We selected target genes based on the GWAS of the CARDIoGRAMplusC4D Consortium7, which identified 47 CAD loci including 63 genes. We added 3 genes located at two East Asian specific loci from Han-Chinese GWAS6. In addition, we selected 9 genes in linkage disequilibrium (r2 > 0.5) with top SNPs in these GWASs according to the 1000 Genomes Projects38 Phase 3 data in studied populations. Furthermore, we added 8 genes that might have a relationship with the susceptibility to atherosclerosis based on an expression quantitative trait locus analysis of mouse and human vascular cells39. Consequently, we selected 83 genes located at 49 loci.
To search for variants with clinical implications, we selected genes satisfying one of the following criteria: (1) there was an established assay for measurement of encoded proteins or genes had been known to be “druggable”40 and (2) the gene-deficient mouse models recapitulated CAD/MI-related phenotypes. Finally, we selected 36 genes located at 19 CAD susceptibility loci (Supplementary Table S1).
Library preparation
We performed multiplex PCR-based targeted sequencing41. Coding DNA sequences (CDS) for 36 targeted genes were defined according to the Consensus CDS (CCDS) database release 1542. Long CDS were divided into 180 base pairs (bps) fragments. We designed, tested and optimized PCR primers for a total of 690 short fragments. Targeted regions were then amplified by the parallel multiplex PCR using a Platinum Multiplex PCR Master Mix (Life Technologies, Carlsbad, CA, USA). After PCR reaction, adaptors with embedded unique 8 base index sequences were ligated to PCR products using KAPA Library Amplification Kit (KAPA Biosystems, Wilmington, MA, USA). Libraries were purified with magnetic beads (AMPure XP), and quantified using a 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA) and qPCR assay (KAPA Biosystems). We used the HiSeq 2500 v2 cluster chemistry (Illumina, San Diego, CA, USA) as a sequencing platform.
Read mapping and variant analysis
Sequencing data were processed with bcl2fastq (version 1.8.4) and converted to fastq files. PCR primer sequences were removed with cutadapt43 (version 1.8). Next, the sequences were aligned to a human genome reference (hg19) using a Burrows-Wheeler Aligner44 (BWA, version 0.7.5). Aligned read files (Sequence Alignment/Map format: sam) were binarized (bam), indexed with SAMTools45 (version 0.1.19), and processed using a Genome Analysis ToolKit46 (GATK version 3.2.2). Reads were locally realigned by GATK IndelRealigner, and variant detection was performed by both a UnifiedGenotyper and HaplotypeCaller, separately. In the HaplotypeCaller process, multiple bam files from the same sample were called simultaneously using “-ERC GVCF” mode, and then all files were jointly genotyped by GATK GenotypeGVCFs. In the UnifiedGenotyper process, a hard filter was applied by GATK VariantFiltration using the following filter parameters: FisherStrand >40.0, QualByDepth <2.0, RMSMappingQuality <40.0 and MappingQualityRankSumTest <−4.0. Finally, two outputs from different callers were merged into one variant call format (VCF) file, and the original bam files were then genotyped again using this merged VCF file by UnifiedGenotyper in GENOTYPE_GIVEN_ALLELES mode.
Quality Control
In addition to the hard filtration mentioned above, variants were excluded if the DP for any base was less than 20 or had a missing rate of >0.05 or a Hardy-Weinberg equilibrium (HWE) P value of <1 × 10−5 in controls. To filter out further false-positive variants, we created histograms of non-reference allele frequency for all called variants and checked by visual inspection whether each histogram consisted of 3 distinct clusters with its peaks at 0, 0.5 and 1.0. Variants that showed abnormal histogram patterns (e.g., continuous distribution) were excluded. Sample-level quality control measures were also performed. Samples with a ratio of mapped reads to total reads less than 0.6 or samples that could not achieve a minimum of 20-fold coverage for at least 95% of the targeted bases were excluded.
To estimate the accuracy of our method, we sequenced 76 Hapmap Japanese-Han Chinese samples. Comparing the variant calls with the 1000 Genomes Project38 Phase 3 data (1000 g), we found 188 variants, while 1000 g samples had 185 in the same targeted region. Among these, 184 were called in both, 1 was found only in 1000 g, and 4 were found only in our sample. This finding implies that our analysis has a sensitivity of 99.5% (184/185) and a positive predictive value of 97.9% (184/188). Furthermore, we assessed the concordance of genotyped data using 2,360 overlapping samples genotyped by Illumina Human610-Quad Beadchips in our previous GWAS36. The concordance between our analysis and the chip data using 187 shared variants was 99.94% (440,765/441,051).
Variant annotation
Variants were annotated with SnpEff47 using the GRCh37.75 database. For variants with different annotations due to multiple transcripts of the gene, the highest impact effect for each variant was selected.
Statistical analyses
We performed single-variant association analysis by 2 × 2 Fisher’s exact test using Plink software48 (Version 1.07). A meta-analysis for the two stages was performed with the Cochran-Mantel-Haenszel (CMH) method. We set the study-wide significance threshold for single-variant association test to 3.1 × 10−5, based on a Bonferroni correction for 1,630 QC passed variants in the discovery stage.
For gene-based association analysis, we employed SKAT17 and CAST16. We categorized variants into three sets of variants based on multiple protein function prediction algorithms: (1) all non-synonymous variants; (2) damaging, defined by all disruptive variants and missense variants annotated as deleterious by all five protein function prediction algorithms, PolyPhen-2 HumDiv, Polyphen2-HumVar, SIFT, MutationTaster and LRT score; and (3) disruptive (null) variants. Although both gain-of-function and loss-of-function variants exist, protein function prediction algorithms were essentially better at predicting loss-of-function variants than gain-of-function variants49. Thus, we employed a two-sided SKAT, which is good at detecting bidirectional effects, in the analysis of variants for the analysis of the 1st category (all non-synonymous variants). We applied a one-sided CAST to the analyses of variants in the 2nd and 3rd categories (damaging and disruptive). Meta-analyses of the two stages were performed using the MetaSKAT R package for SKAT results and the Metafor R package for CAST results. We set the study-wide significance threshold to 4.6 × 10−4, based on a Bonferroni correction for the targeted 36 genes and 3 categories.
To determine the clinical impact of variants in LDLR and PCSK9, we examined LDL cholesterol levels and onset age of MI in the following groups: carriers of LDLR rare variants in each category (all non-synonymous, damaging and disruptive) and carriers of PCSK9 E32K, R93C and disruptive variants. Effects were estimated using multiple linear regression models adjusted for age, gender, BMI, smoking status and cholesterol lowering medications in assessing LDL cholesterol levels, controlling for the same parameters except for age in assessing the onset age of MI. These statistical calculations were performed using R software.
Data availability
All data generated or analysed during this study are included in this article (and its Supplementary Information file).
References
Ruff, C. T. & Braunwald, E. The evolving epidemiology of acute coronary syndromes. Nat Rev Cardiol 8, 140–147, https://doi.org/10.1038/nrcardio.2010.199 (2011).
Marenberg, M. E., Risch, N., Berkman, L. F., Floderus, B. & de Faire, U. Genetic susceptibility to death from coronary heart disease in a study of twins. N Engl J Med 330, 1041–1046, https://doi.org/10.1056/nejm199404143301503 (1994).
Samani, N. J. et al. Genomewide association analysis of coronary artery disease. N Engl J Med 357, 443–453, https://doi.org/10.1056/NEJMoa072366 (2007).
Kathiresan, S. et al. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat Genet 41, 334–341, https://doi.org/10.1038/ng.327 (2009).
Schunkert, H. et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet 43, 333–338, https://doi.org/10.1038/ng.784 (2011).
Lu, X. et al. Genome-wide association study in Han Chinese identifies four new susceptibility loci for coronary artery disease. Nat Genet 44, 890–894, https://doi.org/10.1038/ng.2337 (2012).
Deloukas, P. et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet 45, 25–33, https://doi.org/10.1038/ng.2480 (2013).
van der Harst, P. & Verweij, N. Identification of 64 novel genetic loci provides an expanded view on the genetic architecture of coronary artery disease. Circ Res 122, 433–443, https://doi.org/10.1161/CIRCRESAHA.117.312086 (2018).
Jørgensen, A. B., Frikke-Schmidt, R., Nordestgaard, B. G. & Tybjærg-Hansen, A. Loss-of-function mutations in APOC3 and risk of ischemic vascular disease. N Engl J Med 371, 32–41, https://doi.org/10.1056/NEJMoa1308027 (2014).
Dewey, F. E. et al. Inactivating variants in ANGPTL4 and risk of coronary artery disease. N Engl J Med 374, 1123–1133, https://doi.org/10.1056/NEJMoa1510926 (2016).
Stitziel, N. O. et al. Coding variation in ANGPTL4, LPL, and SVEP1 and the risk of coronary disease. N Engl J Med 374, 1134–1144, https://doi.org/10.1056/NEJMoa1507652 (2016).
Tang, C. S. et al. Exome-wide association analysis reveals novel coding sequence variants associated with lipid traits in Chinese. Nat Commun 6, 10206, https://doi.org/10.1038/ncomms10206 (2015).
Peloso, G. M. et al. Association of low-frequency and rare coding-sequence variants with blood lipids and coronary heart disease in 56,000 whites and blacks. Am J Hum Genet 94, 223–232, https://doi.org/10.1016/j.ajhg.2014.01.009 (2014).
Liu, D. J. et al. Meta-analysis of gene-level tests for rare variant association. Nat Genet 46, 200–204, https://doi.org/10.1038/ng.2852 (2014).
Patel, A. P. et al. Targeted exonic sequencing of GWAS loci in the high extremes of the plasma lipids distribution. Atherosclerosis 250, 63–68, https://doi.org/10.1016/j.atherosclerosis.2016.04.011 (2016).
Morgenthaler, S. & Thilly, W. G. A strategy to discover genes that carry multi-allelic or mono-allelic risk for common diseases: a cohort allelic sums test (CAST). Mutat Res 615, 28–56, https://doi.org/10.1016/j.mrfmmm.2006.09.003 (2007).
Wu, M. C. et al. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet 89, 82–93, https://doi.org/10.1016/j.ajhg.2011.05.029 (2011).
Adzhubei, I. A. et al. A method and server for predicting damaging missense mutations. Nat Methods 7, 248–249, https://doi.org/10.1038/nmeth0410-248 (2010).
Ng, P. C. & Henikoff, S. Predicting deleterious amino acid substitutions. Genome Res 11, 863–874, https://doi.org/10.1101/gr.176601 (2001).
Schwarz, J. M., Cooper, D. N., Schuelke, M. & Seelow, D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods 11, 361–362, https://doi.org/10.1038/nmeth.2890 (2014).
Chun, S. & Fay, J. C. Identification of deleterious mutations within three human genomes. Genome Res 19, 1553–1561, https://doi.org/10.1101/gr.092619.109 (2009).
Fokkema, I. F., den Dunnen, J. T. & Taschner, P. E. LOVD: easy creation of a locus-specific sequence variation database using an “LSDB-in-a-box” approach. Hum Mutat 26, 63–68, https://doi.org/10.1002/humu.20201 (2005).
Leigh, S. E., Foster, A. H., Whittall, R. A., Hubbart, C. S. & Humphries, S. E. Update and analysis of the University College London low density lipoprotein receptor familial hypercholesterolemia database. Ann Hum Genet 72, 485–498, https://doi.org/10.1111/j.1469-1809.2008.00436.x (2008).
Leigh, S. E., Leren, T. P. & Humphries, S. E. Commentary PCSK9 variants: A new database. Atherosclerosis 203, 32–33, https://doi.org/10.1016/j.atherosclerosis.2009.02.006 (2009).
Do, R. et al. Exome sequencing identifies rare LDLR and APOA5 alleles conferring risk for myocardial infarction. Nature 518, 102–106, https://doi.org/10.1038/nature13917 (2015).
Peloso, G. M. et al. Association of Exome Sequences With Cardiovascular Traits Among Blacks in the Jackson Heart Study. Circ Cardiovasc Genet 9, 368–374, https://doi.org/10.1161/CIRCGENETICS.116.001410 (2016).
Helgadottir, A. et al. Variants with large effects on blood lipids and the role of cholesterol and triglycerides in coronary disease. Nat Genet 48, 634–639, https://doi.org/10.1038/ng.3561 (2016).
Cohen, J. et al. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat Genet 37, 161–165, https://doi.org/10.1038/ng1509 (2005).
Abifadel, M. et al. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat Genet 34, 154–156, https://doi.org/10.1038/ng1161 (2003).
Miyake, Y. et al. Genetic variants in PCSK9 in the Japanese population: rare genetic variants in PCSK9 might collectively contribute to plasma LDL cholesterol levels in the general population. Atherosclerosis 196, 29–36, https://doi.org/10.1016/j.atherosclerosis.2006.12.035 (2008).
Versmissen, J. et al. Efficacy of statins in familial hypercholesterolaemia: a long term cohort study. BMJ 337, a2423 (2008).
Nikpay, M. et al. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet 47, 1121–1130, https://doi.org/10.1038/ng.3396 (2015).
Wald, D. S. et al. Child-Parent Familial Hypercholesterolemia Screening in Primary Care. N Engl J Med 375, 1628–1637, https://doi.org/10.1056/NEJMoa1602777 (2016).
Hirata, M. et al. Cross-sectional analysis of BioBank Japan clinical data: A large cohort of 200,000 patients with 47 common diseases. J Epidemiol 27, S9–S21, https://doi.org/10.1016/j.je.2016.12.003 (2017).
Nagai, A. et al. Overview of the BioBank Japan Project: Study design and profile. J Epidemiol 27, S2–S8, https://doi.org/10.1016/j.je.2016.12.005 (2017).
Hirokawa, M. et al. A genome-wide association study identifies PLCL2 and AP3D1-DOT1L-SF3A2 as new susceptibility loci for myocardial infarction in Japanese. Eur J Hum Genet 23, 374–380, https://doi.org/10.1038/ejhg.2014.110 (2015).
Hirota, T. et al. Genome-wide association study identifies eight new susceptibility loci for atopic dermatitis in the Japanese population. Nat Genet 44, 1222–1226, https://doi.org/10.1038/ng.2438 (2012).
Abecasis, G. R. et al. A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073, https://doi.org/10.1038/nature09534 (2010).
Erbilgin, A. et al. Identification of CAD candidate genes in GWAS loci and their expression in vascular cells. J Lipid Res 54, 1894–1905, https://doi.org/10.1194/jlr.M037085 (2013).
Griffith, M. et al. DGIdb: mining the druggable genome. Nat Methods 10, 1209–1210, https://doi.org/10.1038/nmeth.2689 (2013).
Momozawa, Y. et al. Low-frequency coding variants in CETP and CFB are associated with susceptibility of exudative age-related macular degeneration in the Japanese population. Hum Mol Genet 25, 5027–5034, https://doi.org/10.1093/hmg/ddw335 (2016).
Pruitt, K. D. et al. The consensus coding sequence (CCDS) project: Identifying a common protein-coding gene set for the human and mouse genomes. Genome Res 19, 1316–1323, https://doi.org/10.1101/gr.080531.108 (2009).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. journal 17, 10–12, https://doi.org/10.14806/ej.17.1.200 (2011).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760, https://doi.org/10.1093/bioinformatics/btp324 (2009).
Li, H. et al. The sequence slignment/map format and SAMtools. Bioinformatics 25, 2078–2079, https://doi.org/10.1093/bioinformatics/btp352 (2009).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20, 1297–1303, https://doi.org/10.1101/gr.107524.110 (2010).
Cingolani, P. et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strainw1118; iso-2; iso-3. Fly (Austin) 6, 80–92, https://doi.org/10.4161/fly.19695 (2012).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81, 559–575, https://doi.org/10.1086/519795 (2007).
Flanagan, S. E., Patch, A. M. & Ellard, S. Using SIFT and PolyPhen to predict loss-of-function and gain-of-function mutations. Genet Test Mol Biomarkers 14, 533–537, https://doi.org/10.1089/gtmb.2010.0036 (2010).
Acknowledgements
We would like to express our gratitude to Kyota Ashikawa for the HiSeq operation and practical advice on laboratory experiments. Drs Toru Suzuki and Kenichi Aizawa (Center for Epidemiology and Preventive Medicine, The University of Tokyo Hospital) helped to collect the control samples from the University of Tokyo Hospital. We are grateful to the members of BioBank Japan and the Rotary Club of Osaka-Midosuji District 2660 Rotary International in Japan for supporting our study. This work was conducted as a part of the BioBank Japan Project, supported by the Ministry of Education, Culture, Sports, Science and Technology, Japan.
Author information
Authors and Affiliations
Contributions
T.T., H.M., K.I. and Y.M. wrote the manuscript. T.T and K.I. conducted the data analyses. T.T. and Y.M. conducted genotyping. H.M., T.Y. and M.K. collected the samples. H.M., M.K., I.K. and Y.M. designed the study.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Tajima, T., Morita, H., Ito, K. et al. Blood lipid-related low-frequency variants in LDLR and PCSK9 are associated with onset age and risk of myocardial infarction in Japanese. Sci Rep 8, 8107 (2018). https://doi.org/10.1038/s41598-018-26453-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-26453-x
This article is cited by
-
Genome-wide analyses of early-onset acute myocardial infarction identify 29 novel loci by whole genome sequencing
Human Genetics (2023)
-
MGeND: an integrated database for Japanese clinical and genomic information
Human Genome Variation (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.