Abstract
Approximately 10–20% of patients with primary biliary cholangitis (PBC) progress to jaundice stage regardless of treatment with ursodeoxycholic acid and bezafibrate. In this study, we performed a GWAS and a replication study to identify genetic variants associated with jaundice-stage progression in PBC using a total of 1,375 patients (1,202 early-stage and 173 jaundice-stage) in a Japanese population. SNP rs13720, which is located in the 3′UTR of cathepsin Z (CTSZ), showed the strongest association (odds ratio [OR] = 2.15, P = 7.62 × 10−7) with progression to jaundice stage in GWAS. High-density association mapping at the CTSZ and negative elongation factor complex member C/D (NELFCD) loci, which are located within a strong linkage disequilibrium (LD) block, revealed that an intronic SNP of CTSZ, rs163800, was significantly associated with jaundice-stage progression (OR = 2.16, P = 8.57 × 10−8). In addition, eQTL analysis and in silico functional analysis indicated that genotypes of rs163800 or variants in strong LD with rs163800 influence expression levels of both NELFCD and CTSZ mRNA. The present novel findings will contribute to dissect the mechanism of PBC progression and also to facilitate the development of therapies for PBC patients who are resistant to current therapies.
Similar content being viewed by others
Introduction
Primary biliary cholangitis (PBC, MIM 109720), previously called primary biliary cirrhosis, is a chronic autoimmune liver disease characterized by the destruction of intrahepatic bile ducts and progressive cholestasis, leading to liver cirrhosis, jaundice, and hepatic failure1. Although treatment with ursodeoxycholic acid (UDCA), either as monotherapy or in combination with bezafibrate, is very effective for normalization of liver functions and prevention of PBC progression2,3, approximately 10–20% of PBC patients are resistant to these treatments and ultimately progress to hepatic failure4,5,6. However, the mechanisms underlying the progression of PBC are poorly understood, and the existence of two different types of PBC progression, jaundice type and portal hypertension type, has been proposed4.
The high concordance rate of PBC in monozygotic twins in comparison with dizygotic twins, along with the familial clustering of PBC patients, indicates that strong genetic factors are involved in the development of PBC7. Indeed, genome-wide association studies (GWASs) and subsequent meta-analyses have identified susceptibility loci for PBC, including Human Leukocyte Antigen (HLA) and more than 30 non-HLA loci, in individuals of European descent8,9,10,11,12,13,14,15. In addition, GWASs in the Japanese population identified three novel PBC susceptibility loci, including tumor necrosis factor superfamily member 15 (TNFSF15), POU domain class 2 associating factor 1 (POU2AF1), and protein kinase C beta (PRKCB), which have not been identified in European descent16,17. The IL21, IL4R/IL21R, CD28/CTLA4/ICOS, CD58, ARID3A, IL16, and CSNK2A2/CCDC113 loci were also identified as novel susceptibility loci for PBC in Han Chinese subjects18. These GWASs indicate that similar autoimmune pathways of dendritic cell, T-cell, and B-cell activation and/or differentiation, including MAPK-, phosphatidylinositol-, TNF superfamily-, and NFκB-signaling, contribute to the development of PBC in all populations, although the specific PBC susceptibility genes differ somewhat among Europeans and East Asians.
Several genetic loci, including MDR3, ITGAV, CTLA4, CYP7A1, HNF4A, and PPARGC1A, have been reported to be associated with progression to jaundice stage and/or hepatic failure in PBC19,20,21,22. However, these results were obtained by candidate gene approaches that were insufficiently robust. Previously, genetic variants associated with progression in PBC had not yet been identified by whole-genome approaches. Therefore, we performed a GWAS to identify genetic variants associated with jaundice-stage progression in a Japanese population.
Results
GWAS to identify genetic factors associated with disease progression in PBC
Of 1,375 Japanese PBC patients enrolled in this study, 173 were diagnosed as late stage with jaundice (T.bil ≥ 2 mg/dL) regardless of treatment with UDCA or UDCA plus bezafibrate, whereas the remaining 1,202 patients were diagnosed as early stage at the end of observation (Table 1). For the GWAS, we genotyped 1,125 samples (including 150 samples from jaundice-stage patients and 975 from early-stage patients) using the Affymetrix Axiom Genome-Wide ASI 1 Array. No samples were excluded by Dish QC (threshold ≥0.82 for each sample). Genotype calls of 1,125 samples for approximately 600,000 SNPs were determined using the Genotype Console v4.1 software; no samples were excluded based on overall call rate (threshold >0.97). Among the 1,125 samples, no related samples were identified in identity-by-descent testing. Moreover, principal component analysis using 1,125 samples and HapMap samples (43 JPT, 40 CHB, 91 YRI, and 91 CEU samples) showed a clear cluster of studied samples overlapping only with HapMap JPT (Supplementary Figure 1); ultimately, the average overall call rates of 150 jaundice-stage PBC patients and 975 early-stage PBC patients were 99.37% (97.47–99.80) and 99.41% (97.32–99.81), respectively.
For SNP quality control, we applied the following thresholds during data cleaning (Supplementary Table 1): Hardy-Weinberg equilibrium P > 0.001, SNP call rate >95%, and MAF >5% in both early- and jaundice-stage PBC patients. All cluster plots for SNPs with P < 0.0001 from a chi-square test of the allelic model were checked by visual inspection, and SNPs with ambiguous genotype calls were excluded. Of the SNPs on autosomal chromosomes, a total of 426,245 SNPs passed the quality control filters and were used for the association analysis.
A quantile–quantile plot of the distribution of test statistics for the comparison of allele frequencies in early- and jaundice-stage PBC patients revealed that the inflation factor lambda was 1.014 for all tested SNPs (Supplementary Figure 2). Figure 1 shows a genome-wide view of the single-point association data based on allele frequencies in a comparison between 150 jaundice-stage and 975 early-stage PBC patients. Table 2 shows 12 SNPs with P < 5 × 10−5 in the GWAS. The SNP rs13720 (odds ratio [OR] = 2.15, 95% confidence interval [CI] = 1.58–2.94, P = 7.62 × 10−7), which is located in the 3′UTR of cathepsin Z (CTSZ), showed the strongest association with progression to jaundice stage in PBC.
Validation analysis and high-density association mapping
To validate the associations of the SNPs identified by the GWAS, we performed association tests of the 12 candidate SNPs using a total of 1,375 samples (173 jaundice-stage and 1,202 early-stage PBC patients), including the GWAS set of 1,125 samples and a replication set of 250 samples (23 jaundice-stage and 227 early-stage PBC patients) (Table 1). Although none of the 12 SNPs reached genome-wide significance (P = 1.17 × 10−7) in this validation analysis, SNP rs13720 exhibited a borderline significant association with progression to jaundice stage (OR = 2.15, 95% CI = 1.61–2.87, P = 1.36 × 10−7, Table 2).
SNP rs13720 is located in a large linkage disequilibrium (LD) block including negative elongation factor complex member C/D (NELFCD) and CTSZ. Therefore, we performed high-density association mapping using a total of 1,375 samples by selecting 33 tagging SNPs in these loci (Fig. 2). In this analysis, an intronic SNP of CTSZ, rs163800, but not rs13720, exhibited the most significant association with progression to jaundice stage (OR = 2.16, 95% CI = 1.62–2.87, P = 8.57 × 10−8, Supplementary Table 2).
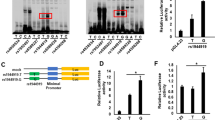
High-density association mapping at NELFCD and CTSZ loci. The top panel shows estimates of pairwise r2 for 33 SNPs used in the high-density association mapping at NELFCD and CTSZ loci (chr 20, nucleotide positions 57514197–57715109, hg19) using a total of 1,375 samples including 173 jaundice-stage and 1,202 early-stage PBC patients. The bottom panel shows P values (●) and OR (○) based on chi-square tests for the allelic model. Red diamond ( ) and red triangle (
) and red triangle ( ) show rs13720 and rs163800, respectively.
) show rs13720 and rs163800, respectively.
In silico analysis of CTSZ and NELFCD genes
Although the top hit, SNP rs163800, was predicted to have minimal binding evidence in the Regulome DB database, four genetic variants (rs3746703, rs151335, rs24048, and rs151336) surrounding rs163800 in strong LD (r2 > 0.8) were predicted to be located in transcriptional regulatory elements, which were identified based on DNase hypersensitivity cluster analyses, prediction of binding sites of transcription factors, and significant associations in eQTL analysis (Supplementary Table 3). Additionally, rs1043219 (located in the 3′UTR of NELFCD) and rs13720 were predicted to have certain biological effects within microRNA binding sites (Supplementary Table 4). Therefore, these polymorphisms, which are in strong LD with rs163800, may affect the gene expression levels of CTSZ and NELFCD.
To assess the biological effects of these variants on mRNA expression, we compared the endogenous expression levels of CTSZ and NELFCD among rs163800 genotypes using the GTEx portal database. Because endogenous CTSZ and NELFCD proteins are abundantly expressed (Supplementary Figure 3), we extracted data from whole blood, transformed fibroblasts, liver, spleen, and EBV-transformed lymphocytes from the GTEx portal database. Among these organs, whole blood and transformed fibroblasts exhibited significant associations between mRNA levels and rs163800 genotype (CTSZ, whole blood: P = 0.0034; CTSZ, transformed fibroblast: P = 0.0075; NELFCD, whole blood: P = 0.0024; NELFCD, transformed fibroblast: P = 7.0 × 10−6, Fig. 3). The risk allele of rs163800 for jaundice-stage progression was associated with elevated NELFCD and reduced CTSZ mRNA levels in both tissue types. Although the associations in liver, spleen, and EBV-transformed lymphocytes did not reach statistical significance, probably due to the small sample size, the patterns in these three tissues resembled those in whole blood and transformed fibroblasts (Supplementary Figure 4). These results suggested that expression of both CTSZ and NELFCD is regulated based on the genotypes of rs163800 or other polymorphisms in strong LD with rs163800.
Endogenous mRNA expression levels of CTSZ and NELFCD as a function of rs163800 genotype in whole blood (A,C) and transformed fibroblast (B,D) Endogenous CTSZ and NELFCD expression data were extracted from the GTEx portal database. Risk (T) and non-risk alleles (C) of rs163800 for jaundice-stage progression in PBC are shown as reference and alteration alleles, respectively. Ref, reference; Het, heterogeneous; Homo, homogenous; Alt, alteration.
Multivariate analysis of anti-nuclear antibodies and CTSZ SNP for jaundice-stage progression in PBC
We previously reported that anti-gp210 and anti-centromere antibodies are risk factors for progression to jaundice stage and late stage without jaundice, respectively, in PBC4. Hence, we performed multivariate analysis (logistic regression analysis) to determine whether the CTSZ SNP is a risk factor independent of anti-nuclear antibodies for jaundice-stage progression in PBC. Consistent with our previous report (4), positivity for anti-gp210 antibodies was a strong risk factor for jaundice-stage progression (OR = 3.04, 95% CI = 2.15–4.31, P = 1.01 × 10−9). In addition, the risk allele of CTSZ SNP, rs163800, was a significant risk factor for jaundice-stage progression (OR = 2.22, 95% CI = 1.63–3.02, P = 8.77 × 10−7), independent of positivity for anti-gp210 antibodies (Table 3).
Discussion
GWAS and subsequent validation studies using a total of 1,375 samples (173 jaundice-stage and 1,202 early-stage Japanese PBC patients) identified a strong association between rs13720, located the NELFCD/CTSZ locus, and progression to jaundice-stage in PBC. In high-density association mapping using selected tagging SNPs from the NELFCD/CTSZ loci, SNP rs163800, located in an intronic region of CTSZ, exhibited a significant association even after Bonferroni correction. This SNP was confirmed as a risk factor for jaundice-stage progression independent of anti-gp210 antibody status. Moreover, in silico functional analysis suggested that expression levels of CTSZ and NELFCD may be influenced by the genotypes of rs163800 or variants in strong LD with rs163800. On the other hand, SNPs in these NELFCD-CTSZ loci did not show any association with the development of PBC (Supplementary Table 5), indicating that the mechanism of PBC-progression to jaundice stage is distinct from that of PBC-development.
Previous studies showed that CTSZ is involved in the development of neurodegenerative disorders, Huntington’s disease and other polyglutamine diseases, multiple sclerosis, and gastric and prostate cancer; furthermore, variants in this gene are associated with susceptibility to tuberculosis23,24,25. In this study, we demonstrated that CTSZ variants are associated with progression to jaundice stage in PBC, mediated through a reduction in CTSZ mRNA levels. CTSZ is a member of the cysteine cathepsins, which are mainly localized in lysosomes, and has exopeptidase activity. CTSZ regulates various cellular physiological functions, including adhesion, migration, invasion, and maturation of immune cells26. Therefore, we postulate that the risk allele of CTSZ might influence the regulation of cellular and/or immune functions through altered cleavage of various molecules, including self and exogenous proteins, leading to progression to jaundice stage in PBC.
NELFCD, which is also referred to as trihydrophobin 1, is an essential component of the NELF complex, which negatively regulates the elongation of transcription by RNA polymerase II27. NELFCD is expressed in a wide range of tissues including heart, brain, lung, liver, kidney, and prostate28. NELFCD inhibits cell migration and cell-cycle progression by negatively regulating MAPK signal transduction29,30. In addition, NELFCD represses estrogen alpha–mediated transactivation and androgen signal transduction, and its expression level is negatively correlated with progression and metastasis of breast cancer28,31,32. A previous report showed that elevated and reduced ER alpha levels in cholangiocytes are associated with aberrant cholangiocyte proliferation in both early- and late-stage PBC and ductopenia in end-stage PBC, respectively33. Collectively, these reports suggest that NELFCD plays a role in progression to jaundice stage in PBC via regulation of MAPK and hormonal signaling pathways.
Several host factors, including proteins encoded by the MDR3, ITGAV, CTLA4, CYP7A1, HNF4A, PPARGC1A, and OCT-1 loci, have been reported to be associated with PBC progression in the Japanese population19,20,21,22. However, these factors were identified by candidate gene approaches, and results are not robust. Among these genes, only one (PPARGC1A) exhibited a significant association (P < 0.05) with disease progression to jaundice stage in our GWAS (Supplementary Table 5). This observation indicates that PPARGC1, a transcriptional activator of CYP7A1, is involved in disease progression of PBC via regulation of bile acid synthesis.
Positivity for anti-gp210 antibodies is a strong risk factor for jaundice-stage progression in PBC4. Recently, the UK-PBC and GLOBE scoring systems were proposed as practical methods to predict long-term outcome of PBC5,6. In these scoring systems, serum levels of biochemical markers such as albumin, bilirubin, alkaline phosphatase, and platelet numbers 1 year after the start of UDCA treatment are used as predictive factors. In a Chinese population, these scoring systems were further optimized by incorporation of anti-gp210 antibody positivity into the predictive factors34. In this study, we identified CTSZ SNP as a novel risk factor, independent of positivity for anti-gp210 antibodies, for jaundice-stage progression in PBC. The scoring system for predicting long-term outcome is currently under investigation by incorporating CTSZ SNP as a novel risk factor in Japanese patients with PBC.
Estimated risk reduction and transplantation by treatment with bezafibrate was reported very recently in patients with PBC by application of UK-PBC and Global PBC risk scores to BEZURO trial (Christophe Corpechot et al. New England Journal of Medicine, 2018 in press). In accordance with this results, the number of PBC patients who progressed to jaundice stage and underwent liver transplantation has been decreasing in Japan after the introduction of bezafibrate for the treatment of PBC, although a large randomized control studies are ongoing to validate the long-term effect of bezafibrate in Japanese PBC patients. Therefore, the role of the risk allele in NELFCD and CTSZ loci for jaundice-stage progression is also an important question to be answered in the ongoing prospective study under treatment with bezafibrate.
In conclusion, our GWAS identified NELFCD and CTSZ as novel loci associated with progression to jaundice stage in PBC. Functional SNPs at these loci may be involved in progression to jaundice stage via alteration of both NELFCD and CTSZ mRNA levels. Although further studies are needed to validate these observations in other independent cohorts, including patients of different ethnicities, our findings will provide novel insights into the genetic architecture of PBC progression to jaundice stage.
Materials and Methods
Ethics statement
All study protocols conform to the relevant ethical guidelines, as reflected in the a priori approval by the ethics committees of Nagasaki Medical Center and all institutes and hospitals that participated in this collaborative study. Written informed consent was obtained from all patients who participated in this study. All samples were anonymized and apart from the respective data. All methods applied in this study were carried out in accordance with the approved guidelines.
Genomic DNA samples and clinical data
Genomic DNA samples from 1,375 Japanese PBC patients were collected by members of the Japan PBC-GWAS Consortium, consisting of 31 hospitals participating in the National Hospital Organization (NHO) Study Group for Liver Disease in Japan (NHOSLJ) and 24 university hospitals participating in gp210 Working Group in Intractable Liver Disease Research Project Team of the Japanese Ministry of Health and Welfare. Patients were diagnosed with PBC if they met at least two of the following three criteria: elevation of cholestatic liver enzymes, positive serum anti-mitochondrial antibodies (AMA), and typical histological findings of liver biopsy specimens. The overlap patients with autoimmune hepatitis was excluded based on the liver biopsy, episodes of high serum titers of ALT ≥200 mIU/ml, and response to prednisolone (0.5 to 1.0 mg/Kg body weight).
The clinical stages for PBC are defined as follows: early stage, no signs indicating portal hypertension or liver cirrhosis; late stage without jaundice, late stage with signs of portal hypertension or liver cirrhosis but without persistent jaundice; jaundice stage, late stage with persistent presence of jaundice, total bilirubin >2 mg/dL. The presence of portal hypertension or liver cirrhosis was diagnosed by ultrasound sonography, computed tomography, and esophago-gastroscopy. In order to efficiently identify genetic factors associated with progression to jaundice stage, we excluded 282 PBC patients in late stage without jaundice at the end of observation; approximately 12.0% of these PBC patients in late stage without jaundice potentially progress to jaundice stage in the future according to our observation that 15 out of 122 PBC patients (12.2%) at late-stage without jaundice progressed to jaundice stage, whereas 15 out of 1028 early-stage PBC patients (1.4%) progressed to jaundice stage and 90 out of 1028 early-stage PBC patients (8.8%) progressed to late stage without jaundice. Thus, of 1,375 Japanese PBC patients, 173 were diagnosed as jaundice-stage, and the remaining 1,202 as early-stage at the end of observation (Table 1).
The 173 jaundice-stage patients had the following clinical characteristics at the end of observation: 153 (88.4%) female; age, 27–76 years (median, 51 years); AMA positivity, 77.9%; glycoprotein (gp)−210 antibody positivity, 58.7%; centromere protein (CENP) antibody positivity, 15.1%; observation period, range 0–25 years, median 6.0 years; treatment, ursodeoxycholic acid only 56.0%, ursodeoxycholic acid + bezafibrate 25.6%, ursodeoxycholic acid + bezafibrate + prednisolone 8.0%. The treatment response one to two years after the initiation of ursodeoxycholic acid or ursodeoxycholic acid + bezafibrare was poor (ALP and/or ALT value > 1.5 upper limit normal) in 35 out of 44 patients (35/44 = 79.5%). For the 1,202 early-stage patients at the end of observation: 1,053 (87.6%) female; age, 23–89 years (median, 60 years); AMA positivity, 84.1%; gp210 antibody positivity, 20.4%; CENP antibody positivity, 23.6%; observation period, range 2–35 years, median 6.0 years; treatment. ursodeoxycholic acid only 68.2.0%, ursodeoxycholic acid + bezafibrate 24.2%, ursodeoxycholic acid + bezafibrate + prednisolone 2.2%. The treatment response one to two years after the initiation of ursodeoxycholic acid or ursodeoxycholic acid + bezafibrare was poor in 59 out of 873 patients (59/873 = 6.8%).
Genomic DNA was extracted from whole peripheral blood of 1,375 Japanese PBC patients using the QIAamp DNA Blood Midi Kit (Qiagen, Tokyo, Japan). One microgram of purified genomic DNA was dissolved in 100 µl of TE buffer (pH 8.0) (Wako, Osaka, Japan), followed by storage at −20 °C until use.
GWAS
Two hundred nanograms of purified genomic DNA from each of 1,125 Japanese PBC patients (including 150 jaundice-stage and 975 early-stage patients) were genotyped as previously described16 using the Affymetrix Axiom Genome-Wide ASI 1 Array. SNP filtering for statistical analysis is summarized in Supplementary Table 1.
Validation analysis and high-density association mapping
To validate the associations of SNPs identified by the GWAS and to carry out high-density association mapping in a genetic region including NELFCD-CTSZ locus, 12 and 33 SNPs, respectively, were genotyped in a total of 1,375 samples using the DigiTag235 and custom TaqMan SNP genotyping assays (Applied Biosystems, Foster City, CA, USA) on a LightCycler 480 Real-time PCR system (Roche, Mannheim, Germany). For the high-density association mapping, 33 tagging SNPs were selected from a region of approximately 200 kb (chr20, nucleotide positions 57514197–57715109, hg19) based on haplotype blocks estimated from Japanese HapMap data using the Haploview software (Table 2).
Databases
Predictions of miRNA binding to CTSZ rs13720 and NELFCD rs1043219 were obtained from FuncPred (https://snpinfo.niehs.nih.gov/snpinfo/snpfunc.html) and PolymiRTS (http://compbio.uthsc.edu/miRSNP/). Predictions of the locations of the variants in transcriptional regulatory elements were obtained from the RegulomeDB database (http://www.regulomedb.org/) and UCSC genome browser (http://genome.ucsc.edu/cgi-bin/hgGateway). Data regarding the correlation between rs163800 genotype and CTSZ or NELFCD expression were obtained from the GTEx portal database (http://gtexportal.org/home/).
Statistical analysis
For the GWAS and validation analysis, P values, ORs, and CIs for the association between SNPs and disease progression in PBC patients were assessed by chi-squared test with a two-by-two contingency table for the allelic model. To avoid false-positive results due to multiple testing, significance levels for the GWAS and validation stages were adjusted based on the number of tested SNPs, i.e., P = 1.17 × 10−7 (0.05/426,245) and P = 1.52 × 10−3 (0.05/33), respectively. Multivariate logistic regression analysis was performed using gp-210 antibody positivity and CENP antibody positivity as covariates.
References
Kaplan, M. M. & Gershwin, M. E. Primary biliary cirrhosis. N Engl J Med 353, 1261–1273 (2005).
Kuiper, E. M. et al. Improved prognosis of patients with primary biliary cirrhosis that have a biochemical response to ursodeoxycholic acid. Gastroenterology 136, 1281–1287 (2009).
Tanaka, A., Hirohara, J., Nakanuma, Y., Tsubouchi, H. & Takikawa, H. Biochemical responses to bezafibrate improve long-term outcome in asymptomatic patients with primary biliary cirrhosis refractory to UDCA. J Gastroenterol 50, 675–682 (2015).
Nakamura, M. et al. Anti-gp210 and anti-centromere antibodies are different risk factors for the progression of primary biliary cirrhosis. Hepatology 45, 118–127 (2007).
Lammers, W. J. et al. Development and Validation of a Scoring System to Predict Outcomes of Patients With Primary Biliary Cirrhosis Receiving Ursodeoxycholic Acid Therapy. Gastroenterology 149, 1804–1812 e1804 (2015).
Carbone, M. et al. The UK-PBC risk scores: Derivation and validation of a scoring system for long-term prediction of end-stage liver disease in primary biliary cholangitis. Hepatology 63, 930–950 (2016).
Selmi, C. et al. Primary biliary cirrhosis in monozygotic and dizygotic twins: genetics, epigenetics, and environment. Gastroenterology 127, 485–492 (2004).
Hirschfield, G. M. et al. Primary biliary cirrhosis associated with HLA, IL12A, and IL12RB2 variants. N Engl J Med 360, 2544–2555 (2009).
Hirschfield, G. M. et al. Variants at IRF5-TNPO3, 17q12-21 and MMEL1 are associated with primary biliary cirrhosis. Nat Genet 42, 655–657 (2010).
Mells, G. F. et al. Genome-wide association study identifies 12 new susceptibility loci for primary biliary cirrhosis. Nat Genet 43, 329–332 (2011).
Liu, X. et al. Genome-wide meta-analyses identify three loci associated with primary biliary cirrhosis. Nat Genet 42, 658–660 (2010).
Hirschfield, G. M. et al. Association of primary biliary cirrhosis with variants in the CLEC16A, SOCS1, SPIB and SIAE immunomodulatory genes. Genes Immun 13, 328–335 (2012).
Liu, J. Z. et al. Dense fine-mapping study identifies new susceptibility loci for primary biliary cirrhosis. Nat Genet 44, 1137–1141 (2012).
Juran, B. D. et al. Immunochip analyses identify a novel risk locus for primary biliary cirrhosis at 13q14, multiple independent associations at four established risk loci and epistasis between 1p31 and 7q32 risk variants. Hum Mol Genet 21, 5209–5221 (2012).
Cordell, H. J. et al. International genome-wide meta-analysis identifies new primary biliary cirrhosis risk loci and targetable pathogenic pathways. Nat Commun 6, 8019 (2015).
Nakamura, M. et al. Genome-wide association study identifies TNFSF15 and POU2AF1 as susceptibility loci for primary biliary cirrhosis in the Japanese population. Am J Hum Genet 91, 721–728 (2012).
Kawashima, M. et al. Genome-wide association studies identify PRKCB as a novel genetic susceptibility locus for primary biliary cholangitis in the Japanese population. Hum Mol Genet 26, 650–659 (2017).
Qiu, F. et al. A genome-wide association study identifies six novel risk loci for primary biliary cholangitis. Nat Commun 8, 14828 (2017).
Ohishi, Y. et al. Single-nucleotide polymorphism analysis of the multidrug resistance protein 3 gene for the detection of clinical progression in Japanese patients with primary biliary cirrhosis. Hepatology 48, 853–862 (2008).
Aiba, Y. et al. Genetic polymorphisms in CTLA4 and SLC4A2 are differentially associated with the pathogenesis of primary biliary cirrhosis in Japanese patients. J Gastroenterol 46, 1203–1212 (2011).
Inamine, T. et al. Association of genes involved in bile acid synthesis with the progression of primary biliary cirrhosis in Japanese patients. J Gastroenterol 48, 1160–1170 (2013).
Inamine, T. et al. A polymorphism in the integrin alphaV subunit gene affects the progression of primary biliary cirrhosis in Japanese patients. J Gastroenterol 46, 676–686 (2011).
Cooke, G. S. et al. Mapping of a novel susceptibility locus suggests a role for MC3R and CTSZ in human tuberculosis. Am J Respir Crit Care Med 178, 203–207 (2008).
Baker, A. R. et al. Genetic susceptibility to tuberculosis associated with cathepsin Z haplotype in a Ugandan household contact study. Hum Immunol 72, 426–430 (2011).
Adams, L. A. et al. Polymorphisms in MC3R promoter and CTSZ 3′UTR are associated with tuberculosis susceptibility. Eur J Hum Genet 19, 676–681 (2011).
Kos, J., Vizin, T., Fonovic, U. P. & Pislar, A. Intracellular signaling by cathepsin X: molecular mechanisms and diagnostic and therapeutic opportunities in cancer. Semin Cancer Biol 31, 76–83 (2015).
Narita, T. et al. Human transcription elongation factor NELF: identification of novel subunits and reconstitution of the functionally active complex. Mol Cell Biol 23, 1863–1873 (2003).
Yang, Y. et al. Trihydrophobin 1 attenuates androgen signal transduction through promoting androgen receptor degradation. J Cell Biochem 109, 1013–1024 (2010).
Cheng, C. et al. Trihydrophobin 1 Interacts with PAK1 and Regulates ERK/MAPK Activation and Cell Migration. J Biol Chem 284, 8786–8796 (2009).
Liu, W. et al. Trihydrophobin 1 is a new negative regulator of A-Raf kinase. J Biol Chem 279, 10167–10175 (2004).
Sun, J. et al. Attenuation of atherogenesis via the anti-inflammatory effects of the selective estrogen receptor beta modulator 8beta-VE2. J Cardiovasc Pharmacol 58, 399–405 (2011).
Zou, W. et al. Negative role of trihydrophobin 1 in breast cancer growth and migration. Cancer Sci 101, 2156–2162 (2010).
Alvaro, D. et al. Estrogen receptors in cholangiocytes and the progression of primary biliary cirrhosis. J Hepatol 41, 905–912 (2004).
Yang, F. et al. The risk predictive values of UK-PBC and GLOBE scoring system in Chinese patients with primary biliary cholangitis: the additional effect of anti-gp210. Aliment Pharmacol Ther 45, 733–743 (2017).
Nishida, N., Mawatari, Y., Sageshima, M. & Tokunaga, K. Highly parallel and short-acting amplification with locus-specific primers to detect single nucleotide polymorphisms by the DigiTag2 assay. PLoS One 7, e29967 (2012).
Acknowledgements
We are indebted to all volunteers who participated in our PBC project. We also thank Drs Toshiki Nikami, Hajime Ota, Hiroshi Kouno, Hirotaka Kouno, Makoto Nakamuta, Nobuyoshi Fukushima, Tatsuji Komatsu, Toshiki Komeda, Shinji Katsushima, Yukio Ohara, Seiji Tsunematsu, Toyokichi Muro, Tsutomu Yamashita, Kaname Yoshizawa, Yoko Nakamura, Masaaki Shimada, Noboru Hirashima, Kazuhiro Sugi, Keisuke Ario, Eiichi Takesaki, Noriaki Naeshiro, Atsushi Naganuma, Hiroshi Mano, Haruhiro Yamashita, Kouki Matsushita, Fujio Makita, Hideo Nishimura, Kiyoshi Furuta, Naohiro Takahashi, Masahiro Kikuchi, Naohiko Masaki, Hitoshi Takaki, Takeaki Sato, Masahiko Takahashi, Tetsuo Yamamoto, Masaharu Koda, Hironori Sakai, Michio Kato, Iwao Yabuuchi, Yuko Nagaoki, Noriaki Naeshiro, Shigeki Hayashi, Koichi Honda, Jinya Ishida, Yukio Watanabe, Masakazu Kobayashi, Michiaki Koga, Takeo Saoshiro, Michiyasu Yagura, Yuji Kamitsukasa, Keisuke Hirata, Yusuke Shimada, Eiji Mita, Kazuhiko Yamauchi, Rie Sugimoto (Members of PBC Research in the NHO Study Group for Liver Disease in Japan (NHOSLJ)), Hajime Takikawa (Department of Medicine, Teikyo University School of Medicine, Tokyo, Japan), Mikio Zeniya (Department of Gastroenterology and Hepatology, Tokyo Jikei University School of Medicine, Tokyo, Japan), Etsuko Hashimoto, Makiko Taniai (Department of Medicine and Gastroenterology, Tokyo Women’s Medical University, Tokyo, Japan), Hiromasa Ohira (Department of Gastroenterology and Rheumatic Diseases, Fukushima Medical University of Medicine, Fukushima, Japan), Takeji Umemura Satoru Joshita (Department of Medicine, Division of Gastroenterology and Hepatology, Shinshu University School of Medicine, Matsumoto, Japan), Kazuhide Yamamoto, Akinobu Takaki (Department of Gastroenterology and Hepatology, Okayama University Graduate School of Medicine, Dentistry and Pharmaceutical Sciences, Okayama, Japan), Masanori Abe, Morikazu Onji (Department of Gastroenterology and Metabology, Ehime University Graduate School of Medicine, Matsuyama, Japan), Kazuhiko Nakao, Tatsuki Ichikawa, Hidetaka Shibata (Department of Gastroenterology and Hepatology, Nagasaki University Graduate School of Biomedical Sciences, Nagasaki, Japan), Satoshi Yamagiwa (Division of Gastroenterology and Hepatology, Niigata University Graduate School of Medical and Dental Sciences, Niigata, Japan), Shuichi Kaneko, Masao Honda, Kuniaki Arai (Department of Gastroenterology, Kanazawa University Graduate School of Medicine, Kanazawa, Japan), Takafumi Ichida, Katsuji Hirano (Department of Gastroenterology and Hepatology, Juntendo University Shizuoka Hospital, Shizuoka, Japan), Masataka Seike (Faculty of Medicine, Oita University, Oita, Japan), Shotaro Sakisaka, Yasuaki Takeyama (Department of Gastroenterology and Medicine, Fukuoka, University School of Medicine, Fukuoka, Japan), Masaru Harada, Michio Senju (The Third Department of Internal Medicine, School of Medicine, University of Occupational and Environmental Health, Kitakyushu, Japan), Osamu Yokosuka, Tatsuo Kanda (Department of Medicine and Clinical Oncology, Graduate School of Medicine, Chiba University, Chiba, Japan), Yoshiyuki Ueno (Department of Gastroenterology, Yamagata University Faculty of Medicine, Yamagata, Japan), Hirotoshi Ebinuma (Division of Gastroenterology and Hepatology, Department of Internal Medicine, Keio Graduate School of Medicine, Tokyo, Japan), Yasuhiko Sugawara, Kiyoshi Hasegawa (Hepatobiliary-Pancreatic Surgery Division, Artificial Organ and Transplantation Division, Department of Surgery, Graduate School of Medicine, The University of Tokyo, Tokyo, Japan), Ken Shirabe, Akinobu Taketomi (Department of Surgery and Science, Kyushu University Graduate School of Medical Sciences, Fukuoka, Japan), Kiyoshi Migita, Masahiro Ito, Shinya Nagaoka, Seigo Abiru, and Hiroshi Yatsuhashi (Clinical Research Center, NHO Nagasaki Medical Center, Omura, Japan), for collecting clinical data and blood samples, and for obtaining informed consent from PBC cases. We are also grateful to Mayumi Ishii, Takayo Tsuchiura (National Center for Global Health and Medicine), Dr. Hiromi Sawai, Megumi Sageshima, Yuko Hirano, Natsumi Baba, Rieko Shirahashi, and Ayumi Ogawa (The University of Tokyo) for technical assistance. This work was supported by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (#23591006, #26293181) to Minoru Nakamura, Yoshihiro Aiba (#15K19357, #17K09449), Yuki Hitomi (#15K19314), and Minae Kawashima (#15K06908); a Grant-in-Aid for Clinical Research from the National Hospital Organization to Minoru Nakamura; a grant from the Research Program for Intractable Disease, provided by the Ministry of Health, Labour and Welfare of Japan to Minoru Nakamura; Grants-in-Aid from the Ministry of Health, Labour and Welfare of Japan to Katsushi Tokunaga (H26-kanenjitsu-kanen-ippan-004) and the Japan Agency for Medical Research and Development to Katsushi Tokunaga, Masao Nagasaki, Yosuke Kawai, Kaname Kojima, and Yuki Hitomi (Platform Program for Promotion of Genome Medicine, 16 km0405205h0101); and grants from The Uehara Memorial Foundation, Takeda Science Foundation, and Senshin Medical Research Foundation to Yuki Hitomi.
Author information
Authors and Affiliations
Contributions
Study concept and design: N.N., Y.H., M.K., M.N.; sample collection: H.N., N.Y., T.T., S.T., A.M., S.Y., Y.S., T.Y., M.T., A.T., K.H., S.S., A.K., S.E., Y.M., S.U., N.K.; genotyping: N.N., Y.A., Y.H., M.K.; statistical analysis: M.K., K.K., Y.K., K.U., Ma. N., K.T.; drafting of the manuscript: Y.A., N.N., M.K., Y.H., K.T., M.N.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Nishida, N., Aiba, Y., Hitomi, Y. et al. NELFCD and CTSZ loci are associated with jaundice-stage progression in primary biliary cholangitis in the Japanese population. Sci Rep 8, 8071 (2018). https://doi.org/10.1038/s41598-018-26369-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-26369-6
This article is cited by
-
SLMO2 is a potential prognostic and immunological biomarker in human pan-cancer
Scientific Reports (2024)
-
Regional heritability mapping identifies several novel loci (STAT4, ULK4, and KCNH5) for primary biliary cholangitis in the Japanese population
European Journal of Human Genetics (2021)
-
Increased expression and altered localization of cathepsin Z are associated with progression to jaundice stage in primary biliary cholangitis
Scientific Reports (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.