Abstract
Human embryonic stem cells (hESCs) exhibit high levels of proteasome activity, an intrinsic characteristic required for their self-renewal, pluripotency and differentiation. However, the mechanisms by which enhanced proteasome activity maintains hESC identity are only partially understood. Besides its essential role for the ability of hESCs to suppress misfolded protein aggregation, we hypothesize that enhanced proteasome activity could also be important to degrade endogenous regulatory factors. Since E3 ubiquitin ligases are responsible for substrate selection, we first define which E3 enzymes are increased in hESCs compared with their differentiated counterparts. Among them, we find HECT-domain E3 ligases such as HERC2 and UBE3A as well as several RING-domain E3s, including UBR7 and RNF181. Systematic characterization of their interactome suggests a link with hESC identity. Moreover, loss of distinct up-regulated E3s triggers significant changes at the transcriptome and proteome level of hESCs. However, these alterations do not dysregulate pluripotency markers and differentiation ability. On the contrary, global proteasome inhibition impairs diverse processes required for hESC identity, including protein synthesis, rRNA maturation, telomere maintenance and glycolytic metabolism. Thus, our data indicate that high proteasome activity is coupled with other determinant biological processes of hESC identity.
Similar content being viewed by others
Introduction
Pluripotent stem cells can replicate indefinitely in an undifferentiated state while retaining their potential to differentiate into all cell lineages1,2. Embryonic stem cells (ESCs) derived from blastocysts are the gold standard of pluripotency. Moreover, somatic cells can be reprogrammed into induced pluripotent stem cells (iPSCs) that share similar characteristics with ESCs3,4. Given their intrinsic abilities, pluripotent stem cells represent an invaluable resource to investigate development and disease, holding great promise for regenerative medicine. As the origin of multicellular organisms, a series of regulatory and quality control mechanisms must operate at high fidelity in these cells5. As such, protein homeostasis (proteostasis) is central for self-renewal, pluripotency and cell fate decisions6,7,8,9.
A key node of the proteostasis network is the ubiquitin proteasome system (UPS), the major selective proteolytic mechanism in eukaryotic cells10,11. Notably, human ESCs (hESCs) and iPSCs have increased proteasome activity compared with their differentiated counterparts12. This enhanced activity is induced by PSMD11/RPN613, a scaffolding subunit that promotes proteasome assembly14. Besides PSMD11, other proteasome regulators (e.g., Psmd14, POMP) are up-regulated in mouse and human pluripotent stem cells6,15,16. Conversely, a mild down-regulation of proteasome activity induces a demise of hESC identity, characterized by decreased mRNA and protein levels of pluripotency markers13,15,17. Concomitantly, pluripotent stem cells with reduced proteasome activity lose their ability to differentiate into neural cells while expressing high levels of markers of endoderm, mesoderm and fibroblast differentiation13.
These findings raise the intriguing question of why ESC function needs enhanced proteasome activity. The proteasome terminates damaged, misfolded and aggregated proteins ensued from stress conditions and misfolding-prone mutations11,18. The accumulation of damaged proteins could alter the immortality of pluripotent stem cells5. Notably, growing evidence indicates that increased proteasome activity prevents the aggregation of damaged proteins such as mutant huntingtin, which underlies Huntington’s disease9,12,13,19. Moreover, the passage of altered proteins to progenitor cells could compromise organismal development and aging. In this regard, enhanced proteasome activity participates in the degradation of damaged proteins in the early steps of mouse ESC differentiation, a key step to generate daughter cells with an intact proteome20,21.
The UPS also modulates half-life of a myriad of regulatory proteins22,23, in a dynamic process adjusted to the specific requirements of a particular cell type and status24,25. Since the differentiation process triggers numerous changes in the cellular proteome, the UPS determines successful organismal development5. In these lines, the proteasome modulates the abundance of key pluripotency factors such as OCT4 or NANOG26,27,28,29. A chain of at least four 48-linked ubiquitins is the primary signal for recognition and degradation by the proteasome30,31. E3 ligases transfer ubiquitin to a target protein, providing specificity to the proteolytic process. Accordingly, over 600 E3 ubiquitin ligases have been identified in humans so far. To prevent proteolysis, deubiqutinating enzymes (DUBs) perform the opposite action. Thus, both E3 and DUBs must operate in a balanced manner to maintain hESC function or activate differentiation26,27. Remarkably, the levels of distinct E3 and DUB enzymes undergo dramatic changes during differentiation of mouse ESCs and cell reprogramming6,9. Recent studies have revealed that distinct proteins with DUB activity such as USP2232,33, USP4434,35 or the proteasome component Psmd146 regulate the levels of core pluripotency transcription factors. Likewise, specific E3 enzymes modulate pluripotency and differentiation29. For instance, Huwe1 polyubiquitinates the pluripotency transcription factor N-myc, promoting its degradation by the proteasome and allowing for neural differentiation of mouse ESCs36.
To obtain further insights into the mechanisms by which increased proteasome activity sustain hESC identity, here we determine the interactome of highly-expressed E3 ligases in these cells (i.e., HERC2, UBE3A or RNF181). Moreover, we perform loss-of-function experiments of these E3s enzymes to examine their impact on hESC identity. Besides its role in the regulation of damaged proteins and endogenous substrates, increased proteasome activity could also be coupled to intrinsic characteristics of pluripotent stem cells. For instance, ESC and iPSCs exhibit up-regulated global translational rates37. To assess this hypothesis, we define the alterations induced by proteasome dysregulation using a shot label-free proteomic approach. Notably, we find changes in key biological processes of hESC identity such as RNA biogenesis, protein synthesis and telomere function, establishing the proteasome as a central regulator of hESCs.
Results
The levels of specific E3 enzymes decrease during differentiation
To obtain insights into the role of the UPS in hESC function, we examined changes in the expression of E3 ubiquitin ligases and DUBs during differentiation. For this purpose, we analysed available proteomics data comparing hESCs with their differentiated neuronal counterparts9 (Supplementary Data 1). We identified 31 DUBs in both hESCs and neurons. Among them, 10 were upregulated and 15 were downregulated during differentiation into neurons. We found that 44 out of 99 identified E3 enzymes increase during neuronal differentiation. In contrast, 35 E3 enzymes were decreased during differentiation into neurons (Supplementary Data 1 and Supplementary Table 1). To identify potential substrates of the UPS in hESCs, we further examined their up-regulated E3 ubiquitin ligases. Our approach was supported by the fact that three of these E3 enzymes were previously reported as regulators of ESC identity (i.e., HUWE136, TRIM3338 and RNF40)34.
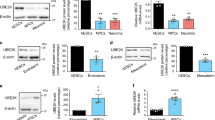
Among the 35 E3 enzymes significantly increased in hESCs, we found 6 ubiquitin ligases with HECT domain while the other E3 enzymes belonged to the RING-domain family39 (Supplementary Table 1). To validate these proteomics data, we performed western blot analysis of representative E3s of both families comparing hESCs with their neural progenitor cell (NPC) and neuronal counterparts. As a control, we also examined the levels of STUB1, an E3 enzyme which slightly increased during neuronal differentiation (Fig. 1a and Supplementary Data 1). At early neural stages, we already observed a downregulation in HECTD1, HERC2, HUWE1, UBE3A, KCMF1, RNF40, RNF181 and UBR7 levels (Fig. 1a). We corroborated the downregulation of distinct E3 ligases during neural differentiation in an independent hESC line (Supplementary Fig. 1). Moreover, these E3 enzymes were also decreased in terminally differentiated neurons validating our proteomics analysis (Fig. 1a and Supplementary Table 1). However, we could not confirm a decrease of TRIM33 levels during neurogenesis as we obtained inconsistent results among independent differentiation experiments (Fig. 1a and Supplementary Fig. 2).
The levels of distinct E3 ubiquitin ligases decrease during differentiation. (a) Western blot analysis of H9 hESCs compared with their neural and neuronal counterparts (i.e., neural precursor cells (NPCs) and terminally differentiated neurons, respectively) with antibodies against distinct E3 ubiquitin ligases. ß-actin is the loading control. E3 enzymes are shown following their classification into HECT and RING-domain families. The images are representative of at least two independent experiments. (b) Quantitative PCR (qPCR) of E3 ubiquitin ligases mRNA levels. Graphs represent the mean ± s.e.m. (relative expression to H9 hESCs) of three independent cells. (c) Western blot analysis of E3 ubiquitin ligases comparing H9 hESCs with their differentiated endoderm precursors counterparts. ß-actin is the loading control. The images are representative of two independent experiments. All cropped blots were run under the same experimental conditions. Uncropped versions of western blots are presented in Supplementary Fig. 13. (d) qPCR analysis of E3 ubiquitin ligases transcript levels upon definitive endodermal differentiation of H9 hESCs. Graphs represent the mean ± s.e.m. (relative expression to H9 hESCs) of four independent cells. (e) mRNA levels of E3 enzymes in H1 hESCs and their differentiated mesoderm and cardiomyocyte counterparts. qPCR data represents the mean ± s.e.m. of three independent experiments. All the statistical comparisons were made by Student’s t-test for unpaired samples. P-value: *(P < 0.05), **(P < 0.01), ***(P < 0.001).
The downregulation in the protein amount of the E3 ubiquitin ligases did not correlate with a reduction in their mRNA levels during neuronal differentiation (Fig. 1b), suggesting a post transcriptional or post-translational regulation of these enzymes. Since multiple E3 ligases are regulated by autoubiquitination rates40, decreased levels of E3s may also indicate their potential activation during differentiation. In hESCs, proteasome inhibition did not result in higher levels of the tested E3 enzymes (Supplementary Fig. 3a). Most importantly, proteasome inhibition did not up-regulate the levels of these E3 enzymes in neurons (Supplementary Fig. 3b). Thus, these data discard a faster turnover of our target E3 enzymes in differentiated cells induced by higher autoubiquination activity.
With the exception of RNF40, the protein levels of most of our target E3 enzymes were also decreased with differentiation into endoderm, indicating that this is not a specific phenomenon associated with the neural lineage (Fig. 1c). Among them, HECTD1, HERC2, HUWE1, UBE3A, RNF181 and TRIM33 were dramatically downregulated while KCMF1 and UBR7 were reduced to a lesser extent (Fig. 1c). In contrast to neuronal differentiation, we also observed a downregulation at the mRNA level (Fig. 1d). Moreover, the gene expression of many of these E3 enzymes (e.g., HECTD1, HERC2, KCMF1) was also downregulated when hESCs differentiated into mesodermal lineages (Fig. 1e). Overall, our results indicate that specific E3 enzymes such as HERC2, UBE3A and RNF181 are highly abundant in hESCs and decrease during differentiation.
HECTD1 interacts with regulators of WNT signaling and glycolysis
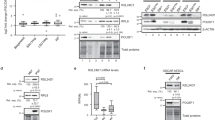
To examine the role of up-regulated E3 enzymes in hESCs, we characterized their interactome. For this purpose, we performed co-immunoprecipitation experiments from hESCs followed by a single shot label-free proteomic approach. Our first target was HECTD1, a HECT E3 ubiquitin ligase involved in the negative regulation of β-catenin (CTNNB1). β-catenin is the main effector of WNT signaling41, one of the regulatory nodes of ESC pluripotency and neural differentiation42,43. We successfully immunoprecipitated significant levels of endogenous HECTD1 (Fig. 2a and Supplementary Fig. 4). Notably, interactome analysis indicated that HECTD1 binds β-catenin and GSK3B, a negative regulator of WNT signaling (Fig. 2b–c and Supplementary Data 2). We also found a strong interaction with subunits of the pyruvate dehydrogenase complex (PDC) (i.e., DLAT, PDHA1, PDHB, PDHX) (Fig. 2b–c). PDC catalyses the mitochondrial conversion of pyruvate into acetyl-CoA, which will be further metabolized through the tricarboxylic acid cycle and electron chain44. The metabolism of ESCs relies on glycolysis rather than mitochondrial respiration for energy production, a process achieved through an inactive PDC complex45. Thus, HECTD1 could participate in maintaining the glycolytic state of hESCs via modulation of PDC activity. In these lines, gene ontology biological process (GOBP) term analysis of the HECTD1 interactome indicated the strongest enrichment for proteins involved in the regulation of Acetyl-CoA biosynthetic process (Fig. 2d and Supplementary Data 2). Interestingly, SIN3A, a transcriptional regulatory protein required for both survival and pluripotency of ESCs46,47, also strongly interacted with HECTD1 (Fig. 2b–c), providing an additional putative link between HECTD1 and ESC function.
HECTD1 interacts with regulators of WNT signaling and glycolysis. (a) Co-immunoprecipitation (co-IP) with HECTD1 and FLAG antibodies in H9 hESCs followed by western blot against HECTD1. We loaded 13.3% of total input and 20% of total immunoprecipitated sample. The images are representative of three independent experiments. All cropped blots were run under the same experimental conditions. The original blots are included in Supplementary Fig. 13. (b) Volcano plot of the interactome of HECTD1 in H9 hESCs (n = 4). Graph represents the −log (p-value) of a two-tailed t-test plotted against the log2 ratio of protein label-free quantification (LFQ) values from co-IP experiments with HECTD1 antibody compared to control co-IP with FLAG antibody. Red colored dots beyond the curved lines indicate some of the most enriched interacting proteins after correction for multiple testing (False Discovery Rate (FDR) adjusted p-value (q-value) <0.2, s0 = 0.1). (c) Scheme indicating the Gene Ontology Molecular Function (GOMF) of HECTD1 interactors (Analysis tool: Cytoscape 3.6.0)137. (d) Bar graph representing the top GOBPs of HECTD1 interactome (P < 0.05) (Analysis tool: PANTHER138 and Gene Ontology Consortium). (e) Venn diagram represents total number and common significant interactors in hESCs, NPCs and neurons (FDR < 0.2 was considered significant, hESCs (n = 4), NPCs (n = 3) and neurons (n = 3).
To determine whether HECTD1 binds these proteins specifically in hESCs, we performed co-immunoprecipitation experiments from NPCs and neurons. Among the 63 HECTD1 interactors identified in hESCs, 39.7% and 28.6% were also significant interactors in NPCs and neurons, respectively (Fig. 2e and Supplementary Data 3). These common interactors included GSK3B, PDC subunits (i.e., DLAT, PDHA1, PDHB, PDHX) and SIN3A in the three cell types tested. Thus, cell-type differences in HECTD1 abundance or activity could determine its regulatory impact on distinct pathways such as WNT signaling or Acetyl-CoA biosynthetic process. Our data also indicate a high percentage of specific interactions of HECTD1 depending on the cell type. Remarkably, 34 proteins interacted with HECTD1 in hESCs while we did not find a significant interaction in NPCs or neurons (Supplementary Data 3). Among these specific interactors, we found β-catenin as well as several regulators of cell cycle and proliferation (e.g., BIRC6, CCNK, CDH1, DDX41, PRPF40A, SRPK2, TAF1), providing a potential link of HECTD1 with the high division rates of these cells. We also identified a hESC-specific interaction of HECTD1 with AURKB, a regulator of telomerase activity in ESCs48. Moreover, HECTD1 specifically interacted in hESCs with proteins involved in gastrulation (i.e., DLD49) and embryonic cell differentiation (e.g., PAXBP150, SRGAP251 and THOC6)52. Thus, our data suggest that cell-specific interactions can also determine endogenous roles of up-regulated E3 enzymes in hESCs.
Knockout of HERC2 alters the transcriptome of hESCs
HERC2 is one of the most up-regulated E3 enzymes in hESCs when compared with their differentiated counterparts (Fig. 1a,c). HERC2 belongs to the HERC gene family, that contains a HECT domain and at least one RCC1-like domain (RLD). HERC2 interacts with chromatin via the RLD domain and participates in DNA repair, DNA replication as well as checkpoint control53. As such, HERC2 is located in a chromosomal region associated with neurodevelopmental disorders, including Prader-Willi and Angelman syndromes54. We immunoprecipitated endogenous HERC2 in hESCs (Fig. 3a and Supplementary Fig. 4) and identified 113 potential interactors (Supplementary Data 4). The three strongest interacting partners were SEC23IP, SRGAP2 and NEURL4 (Supplementary Fig. 5a), which are linked to development51,55,56. These proteins are already reported to interact with HERC2 in human cell lines, validating our data57. In differentiated cells, HERC2 has also been reported to interact with UBE3A, the primary E3 involved in Angelman syndrome58. Moreover, HERC2 has been recently associated with the kinase LRRK2, which plays a role in the familial Parkinson’s disease59. However, we did not observe these interactions in our proteomic data from hESCs, suggesting cell-type differences. On the other hand, the HERC2 interactome was strongly enriched for RNA-binding proteins (Supplementary Data 4). In addition, the most enriched GOBP pathways were related with RNA metabolism (Supplementary Fig. 5b and Supplementary Data 4). The high enrichment for pathways involved in RNA metabolism could indicate indirect interactions mediated by RNA and, therefore, ensuing from immunoprecipitation of RNA-protein complexes rather than direct protein-protein interactions60. To assess whether HERC2 directly interacted with these RBPs, we repeated the interactome analysis treating the samples with RNase A prior to immunoprecipitation with HERC2 antibody60. Whereas the interactions with SEC23IP, SRGAP2 and NEURL4 remained upon RNase treatment, the number of total significant interactors was reduced to 33 proteins (Fig. 3b and Supplementary Data 5). We found a significant interaction with centrosomal proteins (i.e., CENPF, CEP97) as well as RNA-binding proteins such as translation factors (i.e., EIF3A, EIF3C) and the helicase DDX20 (Supplementary Data 5). However, most of the interactions with RNA-binding proteins were lost upon RNase treatment. Moreover, GOBP analysis did not show an enrichment of RNA-related processes among these proteins (Fig. 3c and Supplementary Data 5). These data suggest that the previously identified RNA-binding proteins were immunoprecipitated due to secondary, RNA-mediated interactions.
Knockout of HERC2 does not impair neural differentiation of hESCs. (a) co-IP with HERC2 and FLAG antibodies in H9 hESCs followed by western blot against HERC2. We loaded 13.3% of input and 20% of immunoprecipitated sample. The images are representative of three independent experiments. All cropped blots were run under the same experimental conditions. The original blots are included in Supplementary Fig. 13. (b) Volcano plot of the interactome of HERC2 treating H9 hESC samples with RNase A prior to immunoprecipitation (n = 4). Graph represents the –log (p-value) of a two-tailed t-test plotted against the log2 ratio of LFQ values from co-IP experiments with HERC2 antibody compared to control co-IP with FLAG antibody. Red colored dots indicate some of the most enriched interacting proteins after correction for multiple testing (FDR < 0.2, s0 = 0.1). (c) Bar graph representing the top GOBPs of HERC2 interactome (P < 0.05). (d) Bar graph representing the top GOBPs of the differentially expressed transcripts in HERC2−/− H9 hESCs (P < 0.05). Transcripts showing a log2-fold change at a FDR < 0.05 were retained as significantly differentially expressed. (e) Western blot analysis with antibodies to HERC2, OCT4 and SOX2 comparing wild-type H9 (WT) with HERC2−/− H9 hESCs. ß-actin is the loading control. The images are representative of two independent experiments. All cropped blots were run under the same experimental conditions. The original blots are included in Supplementary Fig. 13. (f) Real-time PCR analysis of pluripotency (upper panel) and germ-layer markers (lower panel) in HERC2−/− H9 hESCs. Graphs (relative expression to wild-type H9 hESCs (WT)) represent the mean ± s.e.m. of four independent experiments. Statistical comparisons were made by Student’s t-test for unpaired samples. P-value: ***(P < 0.001). (g) Immunocytochemistry of H9 hESC with antibodies against OCT4 and SOX2 (markers for pluripotency) and PAX6 (neuroectodermal marker). DAPI staining was used as a marker of nuclei. Scale bar represents 20 μm. (h) Western blot analysis with antibodies to HERC2, NESTIN and PAX6 after 10 days of neural induction. ß-actin is the loading control. The images are representative of three independent experiments. (i) Real-time PCR analysis of pluripotency (upper panel) and neuroectodermal markers (lower panel) in H9 cells after 10 days of neural induction. Graphs (relative expression to WT) represent the mean ± s.e.m. of four independent experiments. Statistical comparisons were made by Student’s t-test for unpaired samples. P-value: *P < 0.05. (j) Immunocytochemistry after 10 days of neural differentiation of HERC2−/− hESCs using antibodies against OCT4, PAX6, NES and SOX1. DAPI was used as a marker of nuclei. Scale bar represents 20 μm.
With the high levels of HERC2 in hESCs and the role of one of its main interactors, SRGAP2, in neurogenesis and spine maturation61, we asked whether HERC2 regulates hESC function or differentiation. For this purpose, we generated a knockout hESC line by CRISPR/Cas9 genome editing (HERC2−/−) and performed transcriptome analysis. We found 1494 transcripts significantly changed in HERC2−/− hESCs (Supplementary Data 6). Interestingly, GOBP analysis indicated strong enrichment for genes involved in the regulation of dendritic spine maintenance and regulation of glial cell proliferation (Fig. 3d and Supplementary Data 6). We also found an alteration in regulators of other developmental process, including nephron and placenta morphogenesis (Fig. 3d). Prompted by these findings, we examined whether these changes in the transcriptome impaired the levels of pluripotency markers. However, HERC2−/− hESCs did not exhibit alterations in the levels of pluripotency markers when compared to control hESCs (Fig. 3e–g), indicating that that the sole deletion of HERC2 does not majorly impact on hESCs function. In addition, we did not find an increase in the expression of markers of the distinct germ layers (Fig. 3f), suggesting that loss of HERC2 does not induce differentiation. We only observe a decrease in the mRNA levels of PAX6, an early marker of neuroectodermal differentiation62 (Fig. 3f). At the pluripotent state, hESCs express marginal amounts of PAX663 making these results difficult to interpret. Nevertheless, a decrease in PAX6 levels at the hESC stage could indicate a dysfunction in their ability to differentiate into NPCs. To assess this hypothesis, we induced neural differentiation. Besides a mild decrease in the mRNA levels of TUJ1, we did not find a significant downregulation in the induction of NPC markers at both protein and transcript levels (Fig. 3h,i). Most importantly, both control and HERC2−/− cultures consisted mostly of PAX6-positive cells at the end of the neural induction treatment (Fig. 3j). Taken together, these results suggest that HERC2 has not a big impact in neural differentiation of hESCs.
Highly abundant E3s interact with stem cell regulators in hESCs
We continued our interactome analysis of up-regulated E3 enzymes focusing on KCMF1, UBR7, UBE3A and RNF181 (Fig. 4a and Supplementary Fig. 4). We excluded HUWE1, TRIM33 and RNF40 as their role and targets have been previously defined in the context of hESC function34,36,38. KCMF1 is a poorly characterized RING finger-domain E3 enzyme64. In Drosophila melanogaster, KCMF1 orthologue regulates MAPK levels through its interaction with UBR465, another E3 ubiquitin ligase. Moreover, the complex KCMF1-UBR4 promotes lysosomal degradation of the E2 ubiquitin protein RAD666. We identified 23 significant KCMF1 interactors in hESCs, including UBR4 (Fig. 4b,c and Supplementary Data 7). One of the most enriched proteins was HMMR (Fig. 4b,c), an important regulator of stemness in murine ESCs through its role in signal transduction at microtubules67. TAF15, another significant KCMF1 interactor, has been previously reported as an epigenetic regulator of ESC pluripotency68. Among the most enriched GOBPs of KCMF1 interactors, we found processes involved in cotranslational protein targeting to the endoplasmic reticulum (Fig. 4d).
Analysis of the interactome of KCMF1 and UBR7 in H9 hESCs. (a) Co-IP with KCMF1, UBR7 and FLAG antibodies in H9 hESCs followed by western blot against the respective E3 ubiquitin ligases. We loaded 13.3% of input and 20% of immunoprecipitated sample. Arrows indicate the specific bands for KCMF1 and UBR7. The images are representative of three independent experiments. All cropped blots were run under the same experimental conditions. The original blots are included in Supplementary Fig. 13. (b) Volcano plot of the KCMF1 interactome (n = 4, FDR <0.2). Red colored dots indicate some of the most enriched interacting proteins after correction for multiple testing. (c) Scheme indicating the Gene Ontology Molecular Function (GOMF) of KCMF1 interactors (d) Bar graph representing the top GOBPs of KCMF1 interactome (P < 0.05). (e) Volcano plot of the UBR7 interactome (n = 4, FDR < 0.2). (f) Scheme indicating the GOMF of UBR7 interactors (g) Bar graph representing the top GOBPs of UBR7 interactome (P < 0.05).
UBR7 belongs to the UBR-box family of E3 ligases. The UBR-box domain contains three zinc finger sites to bind proteins with modified N-terminal amino acid residues based on the N-end rule pathway69. In contrast to other members of the UBR-box family, little is known about UBR7. It was discovered in mammalian sperm, where it was shown to exhibit E3 ligase activity and suggested to be involved in spermiogenesis70. We identified 18 significant interactors of UBR7 in hESCs (Fig. 4e,f and Supplementary Data 8). The most enriched interactor was A2M, a regulator of cytokine availability in the local environment of hESCs and, thus, having a direct impact on pluripotency71 (Fig. 4e,f). Another strong UBR7 interactor was ZSCAN10, which is key for mouse ESC identity72. Similar to HECTD1, UBR7 interacted with distinct PDC subunits (i.e., DLAT, PDHA1 and PDHB) (Supplementary Data 8). Accordingly, GOBP analysis of UBR7 interactors indicated an enrichment in acetyl-CoA biosynthetic pathways (Fig. 4g).
UBE3A, also known as E6-AP, belongs to the family of HECT E3 ubiquitin ligases and was initially discovered as a modulator of p53 degradation73. Genetic abnormalities in the maternally inherited UBE3A are responsible for Angelman syndrome74,75. Conversely, duplication or increased expression of UBE3A is linked to autism spectrum disorders76. UBE3A plays a role in dendritic arborization and synapse maturation77,78,79, as well as cell cycle progression80. Additionally, UBE3A participates in the clearance of several aggregated proteins81,82. Among the UBE3A interactors in hESCs, the most enriched protein was SAE1, an important regulator for reprogramming of human somatic cells83 (Fig. 5a–c and Supplementary Data 9). Another interactor of UBE3A was BCCIP, whose deficiency in mouse leads to impaired neural progenitor self-renewal and differentiation capabilities84. AHNAK, which was also significantly enriched in the UBE3A pull-down, is necessary for proper iPSC generation85. GOBP analysis of UBE3A interactors indicated enrichment for proteins involved in metabolic processes of cellular macromolecules, aromatic and nitrogen compounds as well as mRNAs (Fig. 5d and Supplementary 9). However, we cannot rule out that these interactions with RBPs involved in mRNA stability ensue from indirect RNA-mediated binding and further experiments will be required to asssess direct interaction (e.g., RNase A prior to immunoprecipitation).
Highly abundant E3s interact with stem cell regulators in hESCs. (a) Co-IP with UBE3A, RNF181 and FLAG antibodies in H9 hESCs followed by western blot against the respective E3 ubiquitin ligases. We loaded 13.3% of input and 20% of immunoprecipitated sample. The images are representative of three independent experiments. All cropped blots were run under the same experimental conditions. The original blots are included in Supplementary Fig. 13. (b) Volcano plot of the UBE3A interactome (n = 3, FDR < 0.2). Red colored dots indicate some of the most enriched interacting proteins after correction for multiple testing. (c) Scheme indicating the Gene Ontology Molecular Function (GOMF) of UBE3A interactors (d) Bar graph representing the top GOBPs of UBE3A interactome (P < 0.05). (e) Volcano plot of the RNF181 interactome (n = 4, FDR < 0.2). (f) Scheme indicating the GOMF of RNF181 interactors (g) Bar graph representing the top GOBPs of RNF181 interactome (P < 0.05).
The interactome of RNF181 was highly enriched for subunits of dynactin (specifically, DCTN2, ACTR1A, ACTR1B, DCTN4, DCTN1) (Fig. 5e,f and Supplementary Data 10), a macromolecular complex that regulates not only intracellular transport but also chromosome alignment and spindle organization during cell division86. Dynactin is necessary for proper asymmetric division of embryonic skin progenitors87 and thus may also play a role in the asymmetric divisions invoked by ESCs. Furthermore, RNF181 interacts with DZIP3, an E3 enzyme which regulates developmental genes in mouse ESCs by reorganizing 3D chromatin conformation88. Further experiments will be required to determine whether this interaction is direct or DNA-mediated. Besides the proteasome subunit PSMD3, RNF181 also interacted with distinct subunits of the TRiC/CCT chaperonin complex which is critical for hESC proliferation and differentiation9 (Fig. 5f). The TRiC/CCT complex facilitates the folding of ∼10% of the eukaryotic proteome and mediates protein localization to Cajal bodies, a sub-organelle found in the nucleus of highly proliferative cells, such as hESCs89,90. Accordingly, the most enriched GOBP of RNF181 interactors was the positive regulation of protein localization to Cajal bodies (Fig. 5g and Supplementary Data 10). Prompted by these findings, we examined whether RNF181 also interacts with these proteins in post-mitotic neuronal cells. Among the 46 RNF181 interactors found in hESCs, only 17% were also significant interactors in neurons (Supplementary Fig. 6 and Supplementary Data 11). Although RNF181 bound some of the dynactin subunits in neurons (i.e., DCTN2, ACTR1A, DCTN1), the interaction with DZIP3 as well as TRiC/CCT subunits was lost (Supplementary Fig. 6 and Supplementary Data 11). Taken together, the role of the interacting partners of up-regulated E3 ubiquitin ligases in hESCs indicated that these enzymes might be important for stem cell identity.
Loss of distinct up-regulated E3 enzymes induces changes in the proteome of hESCs
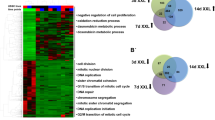
To assess the role of up-regulated E3 ligases in hESC function, we generated stable knockdown (KD) lines for UBR7, UBE3A and RNF181 and analysed their proteome by quantitative proteomics (Supplementary Data 12). We identified the significantly changed proteins upon E3 knockdown and performed GOBP analysis (Supplementary Data 12). Loss of UBR7 changed the levels of 506 proteins, which were enriched for regulators of the generation of energy and metabolites as well as the purine metabolic process (Fig. 6a and Supplementary Data 12). These changes could have an impact on hESCs, which rely on highly active nucleotide synthesis rates for sustaining their rapid proliferation rates and de novo DNA and RNA production91,92. Among the 661 proteins changed upon UBE3A knockdown, GOBP analysis indicated the strongest enrichment for factors involved in the negative regulation of ubiquitin-ligase activity (Fig. 6b and Supplementary Data 12). This is consistent with previous studies, that showed that UBE3A interacts and ubiquitinates several proteasome subunits93,94 and regulates the activity of the proteasome in a ligase-dependent way95,96. Thus, UBE3A could be involved in the regulation of hESC identity through modulation of the proteasome, which is central for maintaining pluripotency13. Loss of RN181 changed the hESC proteome to a lesser extent (202 proteins) compared with UBR7 and UBE3A KD (Supplementary Data 12). Proteins dysregulated in RNF181 KD hESCs were involved mainly in mRNA processing-related pathways (Fig. 6c). Transcription hyperactivity has been proposed as a hallmark of pluripotent stem cells, as the transcription rates markedly decline during differentiation97. In these lines, mRNA-related proteins deregulated in RNF181 KD lines -such as SNRPD1, SRSF5 or HNRNPK- have been proven necessary for stemness in pluripotent cells98,99,100. If E3 interactors are activated for proteasomal degradation by the respective E3 enzyme, we would expect them to increase upon loss of the ubiquitin ligase. However, only a marginal number of the interacting partners were up-regulated upon E3 knockdown despite the numerous changes in the proteome induced by loss of E3 ligases (Fig. 6d).
Loss of distinct up-regulated E3 enzymes induces changes in the hESC proteome. (a) Loss of UBR7 changed the levels of 506 proteins in H9 hESCs (FDR <0.2 was considered significant, n = 3). GOBP analysis (P < 0.05) revealed a strong enrichment for regulators of the purine metabolic process as well as generation of energy and metabolites. (b) Loss of UBE3A changed the levels of 661 proteins in H9 hESCs (FDR < 0.2 was considered significant, n = 3). Bar graph representing the top GOBPs changed upon UBE3A KD (P < 0.05). (c) Loss of RNF181 changed the levels of 202 proteins in H9 hESCs (FDR < 0.2, n = 3). Bar graph representing the top GOBPs changed upon RNF181 KD (P < 0.05). (d) Common proteins between interacting partners of E3 ubiquitin ligases and proteins up-regulated in hESCs with knockdown of the respective E3 enzymes.
One step further was to examine whether loss of UBR7, UBE3A or RNF181 alters hESC identity and differentiation. Loss of UBR7 did not result in significant differences in pluripotency markers compared with control hESCs (Fig. 7a,b). Furthermore, the expression of markers of the distinct germ layers was not impaired (Fig. 7b). We induced neural differentiation of UBR7 KD hESCs and monitored the expression of PAX6. After 10 days of neural induction, PAX6 levels were increased at the same extent in control and UBR7 KD lines (Fig. 7c–e). We also analysed the levels of other neural and neuronal markers and found no differences at this stage (Fig. 7e). To assess whether UBR7 was required for proper differentiation into other germ lineages, we differentiated UBR7 KD hESCs into endoderm (Supplementary Fig. 7). The induction of distinct endoderm markers was similar between control and UBR7 KD lines, suggesting no major role of UBR7 in cell fate decisions towards the endodermal lineage (Supplementary Fig. 7).
Loss of UBR7, UBE3A and RNF181 does not affect the expression of pluripotency markers and neural differentiation. (a) Western blot analysis with antibodies to UBR7 and OCT4 comparing non-targeting shRNA (NT) with UBR7 knockdown (KD) H9 hESCs. We used two independent shRNAs to UBR7 (KD1 and KD2, respectively). ß-actin is the loading control. The images are representative of two independent experiments. All cropped blots were run under the same experimental conditions. The original blots are included in Supplementary Fig. 13. (b) Real-time PCR analysis of pluripotency (left panel) and germ-layer markers (right panel) in UBR7 KD H9 hESCs. Graphs (relative expression to NT shRNA) represent the mean ± s.e.m. of four independent experiments. (c) Western blot analysis after 10 days of neural induction of UBR7 KDH9 hESCs. ß-actin is the loading control. The images are representative of two independent experiments. (d) Immunocytochemistry after 10 days of neural differentiation. OCT4, PAX6, and DAPI staining were used as markers of pluripotency, neuroectodermal differentiation, and nuclei, respectively. Scale bar represents 20 μm. (e) Real-time PCR analysis of pluripotency (upper panel) and neuroectodermal markers (lower panel) in UBR7 KD H9 hESCs after 10 days of neural differentiation. Graphs (relative expression to NT) represent the mean ± s.e.m. of three independent experiments. (f,g) Western blots of UBE3A and OCT4 (f) and RNF181 and OCT4 (g) protein levels. ß-actin is the loading control. The images are representative of two independent experiments. (h) Real-time PCR analysis of pluripotency (upper panel) and germ-layer markers (lower panel) in UBE3A and RNF181 KD H9 hESCs. Graphs (relative expression to NT shRNA) represent the mean ± s.e.m. of three independent experiments. (i) Western blot analysis after 10 days of neural induction of UBE3A and RNF181 KD H9 hESCs. ß-actin is the loading control. The images are representative of two independent experiments. (j) Immunocytochemistry after 10 days of neural differentiation. OCT4, PAX6, and DAPI staining were used as markers of pluripotency, neuroectodermal differentiation, and nuclei, respectively. Scale bar represents 20 μm. All the statistical comparisons were made by Student’s t-test for unpaired samples. P-value: *(P < 0.05), **P < 0.01, ***(P < 0.001), ****(P < 0.0001).
We observed similar results in UBE3A and RNF181 KD hESCs, as pluripotency and differentiation markers remained unchanged after reducing the expression of these E3 enzymes (Fig. 7f–h). We also performed neural (Fig. 7i–j and Supplementary Fig. 8) and endodermal differentiation (Supplementary Fig. 9) from UBE3A and RNF181 KD hESCs and found no differences compared with control cells. Altogether, these results indicate that loss of UBR7, UBE3A or RNF181 alone does not alter the commitment of hESCs to a neuroectodermal or endodermal fate.
Proteasome inhibition induces numerous changes in the proteome of hESCs
To assess whether E3 enzymes potentially regulate proteasomal degradation of specific interactors, we induced acute proteasome dysfunction with the potent MG-132 proteasome inhibitor followed by a shot label-free proteomic approach (Fig. 8a,b). We identified 2735 proteins and hierarchical clustering indicated a clear separation of MG-132-treated hESCs compared with control hESCs based on global protein expression profiles (Supplementary Fig. 10). Quantitative analysis revealed that 899 proteins were significantly changed in MG-132-treated hESCs as compared to control hESCs (Fig. 8b and Supplementary Data 13). Among them, 552 proteins were down-regulated whereas 347 were up-regulated upon proteasome inhibition (Fig. 8b and Supplementary Data 13). Since the latter group could be putative proteasome targets, we integrated these data with the statistically significant E3 interactors. We identified several interactors of each E3 which were also up-regulated upon proteasome inhibitor (Fig. 8c). Among them, we confirmed by western blot the interactions of HECTD1 with EXOC5 and RNF181 with PSMD3 (Fig. 8d). Proteasome inhibition induced a slight increase in both EXOC5 and PSMD3 as we observed by proteomics and western blot (Fig. 8e, Supplementary Fig. 11 and Supplementary Data 13). Thus, a detailed analysis and validation of other E3 interacting partners could reveal novel proteasome targets in hESCs. However, it is also important to note the low number of up-regulated proteins commonly identified in E3 KD lines and proteasome inhibitor-treated hESCs (Fig. 8f). Among the 256 proteins up-regulated in UBR7 KD hESC, only 11 proteins were also increased upon proteasome inhibition (Fig. 8f). Loss of UBE3A resulted in 407 up-regulated proteins. Among them, 22 were also increased upon MG-132 treatment, including TRIOBP and MFGE8, both important for ESC function101,102 (Fig. 8f). Knockdown of RN181 only induced an increase in 4 proteins that were also up-regulated under proteasome inhibition (Fig. 8f). Thus, although a E3-based approach could lead to the discovery of potential proteasomal targets in hESCs, our results indicate that E3s are regulating multiple processes in a proteasome-independent manner that can complicate our analysis. Moreover, we cannot discard compensatory mechanisms or potential functional redundancies among different E3 enzymes103,104,105,106 as suggested by the lack of strong phenotypes in our loss-of-function experiments. In these lines, we have observed multiple common interactors between distinct E3 enzymes (Supplementary Fig. 12).
Proteasome inhibition induces profound changes in the proteome of ESCs. (a) Chymotrypsin-like proteasome activity in H9 hESCs treated with 1 µM MG-132 for 24 h (relative slope to H9 hESCs treated with DMSO). Graph represents the mean ± s.d. of two independent experiments. Statistical comparisons were made by Student’s t-test for unpaired samples. P-value: **(P < 0.01). (b) Volcano plot of LFQ intensities in MG-132-treated H9 hESCs as compared to control DMSO-treated hESCs. The significance of a two-tailed t-test is plotted against the log2 fold change of LFQs (FDR = 0.2, s0 = 1). Proteasome inhibitor: 1 µM MG-132 for 24 h. (c) Common proteins between interacting partners of the E3 ubiquitin ligases and proteins up-regulated in hESCs upon proteasome inhibition. (d) Co-IP with HECTD1, RNF181 and FLAG antibodies in H9 hESCs followed by western blot against EXOC5 and PSMD3. (e) Western blot analysis with antibodies to EXOC5 and PSMD3 of MG-132-treated H9 hESCs. ß-actin is the loading control. The images are representative of three independent experiments. All cropped blots were run under the same experimental conditions. The original blots are included in Supplementary Fig. 13. (f) Common up-regulated proteins between E3 knockdown hESCs and MG-132-treated hESCs. (g) Bar graph representing the top GOBPs of the up-regulated proteins upon proteasome inhibitor treatment (P < 0.05). (h) Bar graph representing the top GOBPs of the down-regulated proteins upon proteasome inhibitor treatment (P < 0.05).
Another potential hypothesis is that enhanced proteasome activity of hESCs is not directly linked to the degradation of endogenous substrates. In this model, increased proteasome activity could be coupled with intrinsic characteristics of pluripotent stem cells. In support of this hypothesis, we observed a higher number of down-regulated proteins than increased proteins under proteasome inhibition (552 down-regulated versus 347 up-regulated). To gain insight into the effects of proteasome dysfunction in hESCs, we performed GOBP term analysis of these altered proteins. Analysis of up-regulated proteins indicated strong enrichment for genes involved in proteasomal-mediated ubiquitin degradation, including numerous proteasome subunits (Fig. 8g and Supplementary Data 13). Among them, we found an up-regulation of the proteasome activator PSMD11/RPN613. Thus, these data suggest that hESCs trigger the expression of proteasome subunits as a compensatory mechanism to ameliorate the proteostasis decline induced by proteasome inhibition. In line with these results, we also found a marked up-regulation of proteins involved in chaperone-mediated protein folding and refolding (e.g., HSPA1A, DNAJC7, DNAJB1, HSPA8, HSPD1 and the stress-induced HSP90AA1), protein folding in the endoplasmic reticulum (ER) and ubiquitin-dependent ER-associated degradation (ERAD) (e.g., ERLIN2, VCP). Moreover, we found GOBP enrichment for regulators of stem cell differentiation, cell cycle and WNT signalling pathway (Fig. 8g and Supplementary Data 13). However, this enrichment was essentially due to increased levels of proteasome subunits, which are well-known regulators of the aforementioned biological processes.
On the other hand, GOBP term analysis of down-regulated proteins upon MG-132 treatment revealed a strong enrichment for factors involved in different pathways linked with hESC maintenance (Fig. 8h and Supplementary Data 13). For instance, proteasome dysfunction induced downregulation of activators of ribosome biogenesis and translation (Fig. 8h), a process that may compensate the decline in proteolytic ability. However, these changes could also trigger differentiation as ESCs rely on enhanced expression of ribosomal subunits and global translational rates to sustain their self-renewal and pluripotency37,107. Moreover, ESCs produce high levels of pre-rRNA108 and we found a down-regulation of proteins involved in maturation of rRNA (TSR1, HEATR1, EMG1) (Fig. 8h). In addition, proteasome dysfunction decreased the levels of distinct modulators of mRNA transport such as the nuclear pore complex protein NUP153, which is required for ESC pluripotency109.
Notably, proteasome inhibition also induced the down-regulation of factors involved in telomere maintenance and organization (e.g., RIF1, NOP10) (Fig. 8h), a key process for hESC maintenance110,111. Besides these changes, we found a decrease in the levels of multiple proteins involved in catabolic carbohydrate process (e.g., the glycogen phosphorylases PYGL/PYGB) as well as generation of precursor metabolites and energy (Fig. 8h). Remarkably, we observed a downregulation of hexokinase II (HK2), which is required for the high glycolytic rates of hESCs92. We also found a decrease in LIN28A (Supplementary Data 13), a core component of hESC identity that regulates the translation and stability of numerous mRNAs112,113,114,115, modulating key biological processes such as metabolism116. Thus, proteasome dysfunction impairs the protein levels of multiple factors involved in biological processes required for hESC function, suggesting a potential link between enhanced proteasome and other determinants of hESC identity such as metabolism, RNA biogenesis and telomere maintenance.
Discussion
Pluripotent stem cells exhibit intrinsic high levels of proteasome activity12. Recent work demonstrates that this enhanced proteasome activity is essential for the striking ability of hESCs to maintain their proteostasis9,12,13,19. Besides this role in protein quality control, here we aimed to uncover additional molecular mechanisms by which proteasome activity regulates hESC function. First, we assessed whether increased proteasome is also required to regulate the concentration of endogenous targets in these cells by identifying highly-expressed E3 enzymes. Then, we characterized the interactome of the up-regulated E3 ligases and integrated these data with our proteomics experiments in E3-knockdown and proteasome inhibitor-treated hESCs. Unexpectedly, we identified a relatively low number of potential E3 substrates which were also increased upon proteasome inhibition. The difficulties in the identification of E3 substrates have been previously shown to be notoriously challenging117,118. First, protein ubiquitination is a highly dynamic process modulated by the coordinated action of E3 ligases and DUBs, which can remove the ubiquitin chains of E3 substrates. The balanced regulation of both competing processes could determine the fate of specific targets depending on the cellular status and environment. In this context, changes in the activity of specific DUBs could compensate the silencing of E3 enzymes. In a further level of complexity, DUBs can directly modulate the activity and turnover of E3 ligases119. Thus, a further understanding of the equilibrium between DUB and E3 activities in proteasomal degradation of pluripotency factors is of central importance. Secondly, the interaction between the E3 ligases and the substrates is rapid and weak. Hence, this transient interaction will make it especially challenging to isolate the complex E3-substrate120. Finally, we believe that significant degrees of redundancy and compensatory mechanisms among the E3 enzymes could be particularly relevant for our study. A substrate may be ubiquitinated by multiple E3s in the same or different lysines and even one single protein might be ubiquitinated by distinct enzymes under different conditions or even within single cellular compartments121. E3 substrates can be modified with a single ubiquitin or chains of ubiquitin at one or multiple sites. This variety in the ubiquitination generates a highly diverse topology of poly-ubiquitin chains, which not always targets the substrate towards degradation through the proteasome, but will have several different functions (i.e., intracellular signalling events)122. Thus, the interacting partners found in our proteomic analysis could not be substrates of the proteasome, but rather have their activity/function modified upon ubiquitination or simply be interacting proteins which might participate in an E3-containing protein complex. Nevertheless, our interactome analysis in hESCs may contribute to the understanding of the role of these E3 ubiquitin ligases.
In order to establish whether the E3 ubiquitin ligases have an impact on hESC identity, we silenced their expression and analysed stem cell pluripotency and differentiation capacities. However, we did not find a major effect on stem cell markers and their differentiation capacity upon loss of single E3 ubiquitin ligases. As mentioned above, that could be explained by potential functional redundancies between E3 ligases, as it has been previously shown for distinct E3s105. One example is the yeast transcriptional repressor alpha2, which can be independently marked for proteasomal degradation by the E3 enzymes Doa10 and Slx5/Slx8106. Moreover, the E3 ligases SAN1 and UBR1 can independently regulate the chaperone-dependant ubiquitination of misfolded proteins in yeast103. In mammalian cells, the tumour suppressor p53 can be degraded through the action of many E3 ligases, such as COP1, WWP1, E4F1 and PIRH2, which provides an additional layer of complexity in the regulation of the UPS104. These redundant mechanisms could ensure proper degradation of a particular substrate, even when one of the E3s might not be functional or overloaded by the number of targets to ubiquitinate. Furthermore, the different E3s might show subtle differences depending on their distinct subcellular localizations to enable intracellular-specific ubiquitination of the same substrate. Given the complex roles of E3s in maintaining cellular homeostasis and development, pluripotent stem cells likely rely on a tight coordination of redundant and compensatory mechanisms to maintain their function. Thus, simultaneous deletion of distinct E3 ligases might be necessary to induce a strong demise in their pluripotency. In addition, we cannot discard that the lack of phenotype of E3-depleted hESCs ensues from the activation of other proteostatic pathways that counterbalance UPS disturbances. For instance, it has been demonstrated that inhibition of proteasome induces autophagy as a compensatory response, in a p62-mediated manner123,124.
In parallel to this approach, we also performed quantitative proteomic analysis of hESCs treated with proteasome inhibitor. By integrating these data with our interactome and loss of function experiments for distinct E3 ligases, we identified potential targets of the proteasome in hESCs. Most importantly, global proteasome inhibition experiments showed that besides its key role in maintaining proteostasis in hESCs, the proteasome is also coupled to other determinant biological processes of hESC identity, such as their intrinsic metabolism, RNA biogenesis and telomere maintenance. Overall, here we provide a comprehensive characterization of the UPS system in hESCs that could have important implications for a further understanding of these cells.
Experimental Procedures
hESCs culture and differentiation
The human H9 (WA09) and H1 (WA01) lines were obtained from WiCell Research Institute. H9 and H1 were maintained on Geltrex (ThermoFisher Scientific) using mTeSR1 (Stem Cell Technologies). Undifferentiated hESCs colonies were passaged using a solution of dispase (2 mg ml−1) and scraping the colonies with a glass pipette. All the cell lines used in this study were tested for mycoplasma contamination at least once every three weeks and no mycoplasma contamination was detected. All research involving hESCs lines was performed with approval of the German Federal competent authority (Robert Koch Institute) and in accordance with the relevant guidelines and regulations.
Neural differentiation of H9 cell line was performed following the monolayer culture method as described previously125 with STEMdiff Neural Induction Medium (Stem Cell Technologies) on polyornithine (15 μg ml−1)/laminin (10 μg ml−1)-coated plates. Undifferentiated hESCs were rinsed once with PBS and then we added 1 ml Gentle Dissociation Reagent (Stem Cell Technologies) for 5 min. After incubation at 5 min, we gently collected hESCs and 2 ml of Dulbecco’s Modified Eagle Medium (DMEM)-F12 + 10 μM ROCK inhibitor (Abcam). Afterward, we centrifuged cells at 300 g for 10 min. Cells were resuspended on STEMdiff Neural Induction Medium + 10 μM ROCK inhibitor and plated on polyornithine (15 μg ml−1)/laminin (10 μg ml−1)-coated plates. For neuronal differentiation, NPC at passage 4 were dissociated with Accutase (Stem Cell Technologies) and plated into neuronal differentiation medium (DMEM/ F12, N2, B27 (ThermoFisher Scientific), 1 mg ml−1 laminin (ThermoFisher Scientific), 20 ng ml−1 brain-derived neurotrophic factor (BDNF) (Peprotech), 20 ng ml−1 glial cell-derived neurotrophic factor (GDNF) (Peprotech), 1 mM dibutyryl-cyclic AMP (Sigma) and 200 nM ascorbic acid (Sigma)) onto polyornithine/laminin-coated plates. Cells were differentiated for 1 month, with weekly feeding of neuronal differentiation medium. Endoderm differentiation of H9 hESCs was performed using STEMdiff Definitive Endoderm Kit (Stem Cell Technologies).
Cardiomyocyte differentiation of H1 hESCs was performed as described in ref.126. Confluent H1 hESCs were dissociated into single cells with Accutase at 37 °C for 10 min followed by inactivation using two volumes of F12/DMEM. Cells were counted and 230,000 cells cm2 where plated in ITS medium (Corning), containing 1.25 mM CHIR 99021 (AxonMedchem) and 1.25 ng ml−1 BMP4 (R&D), and seeded on Matrigel-coated 24-well plates. After 24 h, medium was changed to TS (transferrin/selenium) medium. After 48 h, medium was changed to TS medium supplemented with 10 mM canonical Wnt-Inhibitor IWP-2 (Santa Cruz) for 48 h. Then, medium was changed to fresh TS until beating cells were observed at days 8–10. Finally, medium was changed to Knockout DMEM (ThermoFisher Scientific) supplemented with 2% FCS, L-Glutamine and Penicillin/Streptomycin until cells were used for downstream analysis.
Lentiviral infection of H9 hESCs
Lentivirus (LV)-non-targeting shRNA control, LV-UBR7 shRNA #1 (TRCN0000037025), LV-UBR7 shRNA #2 (TRCN0000037026), LV-UBE3A shRNA #1 (TRCN0000419838), LV-UBE3A shRNA #2 (TRCN0000003368), LV-RNF181 shRNA #1 (TRCN0000364405) and LV-RNF181 shRNA #2 (TRCN0000022389) in pLKO.1-puro vector were obtained from Mission shRNA (Sigma). For generation of stable hESCs transfected lines, H9 hESCs growing on Geltrex were incubated with 10 μM ROCK inhibitor for one hour and then individualized using Accutase. Fifty thousand cells were infected in suspension with 25 μL of concentrated virus in the presence of 10 μM ROCK inhibitor. Cell suspension was centrifuged to remove the virus and plated back on a feeder layer of mitotically inactivated MEF in CDF12 media containing ROCK inhibitor (DMEM/F12 plus 20% Knock-out Serum Replacement, β-mercaptoethanol (0.1%), 0.1 mM non-essential-aminoacids, Glutamax (1%), and 10 ng/mL bFGF). After few days in culture, cells were selected for lentiviral integration with 2 µg ml−1 puromycin (ThermoFisher Scientific) and remaining colonies were manually passaged onto fresh MEFs to establish new hESC lines.
Generation of HERC2 −/− hESCs by CRISPR/Cas9 system and colony analysis
HERC2 genomic sequence was obtained from ENSEMBL Genome browser. Two guide sequences (Guide A, targeting the intron 4–5, F: CCTCAGTTTCTTCATCCATAAAAC, and R: CAGCACTGCTTGACAGTGCTGGG; Guide B, targeting the fifth exon, F: CCGTCCAGTCAGCCACCACCACC, and R: CAGCCCTGCGACTCAAGCAGAGG), were generated using the Zinc Finger Consortium online resource (http://zifit.partners.org/ZiFiT/). Guide-carrying plasmid were designed as previously described in refs127,128 using Cas9-puromycine selection plasmid (pX335-U6-Chimeric_BB-CBh-hSpCas9n(D10A) was a gift from Feng Zhang (Addgene plasmid # 42335)). H9 hESCs were plated on MEF-containing plates and transfected with 1 μg of each of the guide-carrying plasmids using FuGene HD (Promega). 24 h after the transfection, 0.5 μg ml−1 puromycine selection was performed for 24 h followed by maintenance of hESCs with CDF12 media. Single cell split was performed prior to colony pick for genotyping. Genomic DNA isolation was done using QuickExtract (Epicentre). PCR using primer pairs surrounding HERC2 fifth exon was performed (Forward: TAGAGAGAGGCAGTGTGCCA, and Reverse: TGGCTGGCTTCCACAGTTAC) to identify HERC2−/− hESCs.
Immunoprecipitation of HERC2, HECTD1, RNF181, KCMF1 and UBR7 for interaction analysis
hESCs were lysed in RIPA buffer (50 mM Tris-HCl (pH 6.7), 150 mM NaCl, 1% Triton x-100, 1% sodium deoxycholate, 1 mM EDTA, 1 mM PMSF) and supplemented with protease inhibitor (Roche). Lysates were homogenized through syringe needle (27 G) and centrifuged at 13,000 g for 15 min at 4 °C. For RNase A-treated samples, the supernatant was incubated with 125 µl ml−1 RNAse A (ThermoFischer) for one hour on ice. Otherwise, incubation with the antibody was performed. 330 µg of protein-containing samples were incubated for 30 minutes with HERC2 antibody (Abcam, # ab85832, 4 µg) or UBR7 antibody (Thermo Scientific, # PA5-31559, 4 µg) or KCMF1 antibody (Abcam, # ab80287, 4 µg) or RNF181 antibody (Thermo Scientific, PA5-31008, 4 µg) or HECTD1 antibody (Abcam, #ab101992, 4 µg) on ice. As a control, the same amount of protein was incubated with anti-FLAG antibody (SIGMA, F7425, 4 µg). Subsequently samples were incubated with 100 µl of µMACS Micro Beads for 1 h at 4 °C with overhead shaking. After incubation, samples were loaded to precleared µMACS column (# 130-042-701). Beads were washed three times with 50 Mm Tris (pH 7.5), 150 mM NaCl, 5% glycerol, 0.05% Triton and then five times with 50 Mm Tris (pH 7.5) and 150 mMNaCl. For elution of the proteins for further Western Blot analysis, the beads were incubated with 50 µl of pre-heated (95 °C) 1X Laemmli Buffer for 5 minutes and collected into eppendorf tubes. The samples were boiled for 5 minutes at 95 °C and loaded in a SDS-PAGE gel. In Supplementary Fig. 4, we also show flow-through controls, that correspond to the supernatant that remains after centrifuging the tubes containing the antibody as well as the beads. For proteomics analysis, after the washes we performed partial digestion on the beads using 25 µl of 2 M Urea in 25 mM Tris, 7.5 Mm ammonium bicarbonate, 1 mM DTT and 5 ng/ml trypsin, pH = 8.0 for 30 minutes at room temperature. The samples were then eluted twice with 50 µl of 2 M Urea, 25 mM Tris, 7.5 mM ammonium bicarbonate, 5 mM Iodacetamide and left for fully digestion overnight at room temperature in the dark. For stopping the digestion, 5 µl of 100% formic acid was added to samples. Peptides were cleaned up using stage tip extraction129.
Immunoprecipitation of UBE3A for interaction analysis
hESCs were lysed in RIPA buffer (50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% Triton x-100, 1% sodium deoxycholate, 0.1% SDS, 1 mM EDTA, 1 mM PMSF) supplemented with protease inhibitor (Roche). Lysates were homogenized through syringe needle (27 G) and centrifuged at 13,000 g for 15 min at 4 °C. After preclearing the supernatant with Protein A agarose beads (Pierce), the samples were incubated overnight with UBE3A antibody (Cell Signaling, # 7526 1:50) on the overhead shaker at 4 °C. As a control, the same amount of protein was incubated with anti-FLAG antibody (SIGMA, F7425, 1:50) in parallel. Subsequently, samples were incubated with 30 µl of Protein A beads for 1 h at room temperature. After this incubation, samples were centrifuged 5 minutes at 5,000 g at room temperature and the pellet was washed three times with the RIPA buffer. For elution of the proteins for further western blot analysis, the pellet was incubated with pre-heated (95 °C) 2X Laemmli Buffer, boiled for 5 minutes and centrifuged 5 minutes at maximum speed. The supernatant was taken and loaded in a SDS-PAGE gel. For further proteomics analysis, after the washing steps, partial digestion of the proteins was performed on the agarose beads using 25 µl of 2 M Urea in 25 mM Tris, 7.5 mM ammonium bicarbonate, 1 mM DTT and 5 ng/ml trypsin, pH = 8.0 for 30 minutes at room temperature. Then, the samples were eluted 2 times with 50 µl of 2 M Urea, 25 mM Tris, 7.5 mM AmBic 5 mM Iodacetamide and were fully digested overnight at room temperature in the dark. For stopping the digestion, 5 µl of 100% formic acid was added to the samples. Peptides were cleaned up using stage tip extraction129.
Sample preparation for quantitative proteomics and analysis
For analysis of MG132 inhibited cells and hESCs, NPC and neurons we performed tandem mass tag (TMT) proteomics. Cells were collected in 8 M urea buffer, 50 mM ammonium bicarbonate, homogenized with a syringe and cleared using centrifugation (16,000 g, 20 min). Supernatants were reduced by addition of DTT to a final concentration of 5 mM and incubated at room temperature for 30 minutes. Iodoacetamide was added to the samples to a final concentration of 5 mM to alkylate the cysteines and incubated in the dark for 45 minutes. Lys-C (Wako, Richmond, VA) was added for digestion at a ratio 1:100 enzyme/substrate and incubated for three hours at room temperatures. Then, lysates were diluted to 2 M Urea with 50 mM ammonium bicarbonate, after which 10 ng μl−1 Trypsin was added and incubated overnight at room temperature. Trypsin digestion was stopped by addition of formic acid and peptides were cleaned up using stage tip extraction129. The liquid chromatography tandem mass spectrometry (LC-MS/MS) equipment consisted out of an EASY nLC 1000 coupled to the quadrupole based QExactive instrument (Thermo Scientific) via a nano-spray electroionization source. Peptides were separated on an in-house packed 50 cm column (1.9 µm C18 beads, Dr. Maisch) using a binary buffer system: A) 0.1% formic acid and B) 0.1% formic acid in ACN. The content of buffer B was raised from 7% to 23% within 120 min and followed by an increase to 45% within 10 min. Then, within 5 min buffer B fraction was raised to 80% and held for further 5 min after which it was decreased to 5% within 2 min and held there for further 3 min before the next sample was loaded on the column. Eluting peptides were ionized by an applied voltage of 2.2 kV. The capillary temperature was 275 °C and the S-lens RF level was set to 60. MS1 spectra were acquired using a resolution of 70,000 (at 200 m/z), an Automatic Gain Control (AGC) target of 3e6 and a maximum injection time of 20 ms in a scan range of 300–1750 Th. In a data dependent mode, the 10 most intense peaks were selected for isolation and fragmentation in the HCD cell using a normalized collision energy of 25 at an isolation window of 2.1 Th. Dynamic exclusion was enabled and set to 20 s. The MS/MS scan properties were: 17.500 resolution at 200 m/z, an AGC target of 5e5 and a maximum injection time of 60 ms. All label-free proteomics data sets were analysed with the MaxQuant software130 (release 1.5.3.8). We employed the LFQ mode131 and used MaxQuant default settings for protein identification and LFQ quantification. All downstream analyses were carried out on LFQ values with Perseus (v. 1.5.2.4)132.
Quantitative proteomics analysis of E3 enzymes
For the characterization of protein expression differences in E3 enzymes comparing H9 hESCs with their neuronal counterparts, we analysed the quantitative proteomics data published in ref.9. Statistical comparisons were made by Student’s t-test. Discovery Rate (FDR) adjusted p-value (q-value) was calculated using the Benjamini–Hochberg procedure. All the proteins at a FDR level below 0.2 were retained as significantly differentially expressed (Supplementary Data 1). Then, we intersected the human E3 network annotated in133 with this proteomics dataset (Supplementary Table 1).
RNA sequencing
Total RNA was extracted using RNAbee (Tel-Test Inc.). Libraries were prepared using the TruSeq Stranded mRNA Library Prep Kit. Library preparation started with 1 µg total RNA. After selection (using poly-T oligo-attached magnetic beads), mRNA was purified and fragmented using divalent cations under elevated temperature. The RNA fragments underwent reverse transcription using random primers followed by second strand cDNA synthesis with DNA Polymerase I and RNase H. After end repair and A-tailing, indexing adapters were ligated. The products were then purified and amplified (20 µl template, 14 PCR cycles) to create the final cDNA libraries. After library validation and quantification (Agilent 2100 Bioanalyzer), equimolar amounts of library were pooled. The pool was quantified by using the Peqlab KAPA Library Quantification Kit and the Applied Biosystems 7900HT Sequence Detection System. The pool was sequenced on an Illumina HiSeq. 4000 sequencer with a paired- end (2 × 75 bp) protocol. We used the human genome sequence and annotation (EnsEMBL 79) together with the splice-aware STAR read aligner134 (release 2.5.1b) to map and assemble our reference transcriptome. Subsequent transcriptome analyses on differential gene and transcript abundance were carried out with the cufflinks package135.
Western Blot
Cells were scraped from tissue culture plates and lysed in non-denaturing lysis buffer (10 mM Tris-HCL, pH 7.4, 10 mM EDTA, 50 mM NaCl, 50 mM NaF, 1% Triton X-100, 0.1%SDS, supplemented with 2 mM sodium orthovanadate, 1 mM PMSF and protease inhibitor (Roche)) and homogenized through a syringe needle (27 G). Cell lysates were centrifuged at 15,000 g for 15 min at 4 °C and protein concentration was measured from the supernatant using BCA protein assay (Thermo Scientific). Approximately 30 μg of total protein was separated by SDS–PAGE, transferred to PVDF membranes (Millipore) and subjected to immunoblot. Western blot analysis was performed with anti-HERC2 antibody (Abcam, # ab85832 1:2000), anti- UBR7 antibody (Thermo Scientific, # PA5-31559 1:2000), anti- KCMF1 antibody (Abcam, # ab80287, 1:3000), anti-RNF181 antibody (Thermo Scientific, PA5-31008, 1:2000), anti- HECTD1 antibody (Abcam, #ab101992, 1:1000), anti-UBE3A antibody (Cell Signaling, # 7526 1:1000), anti-RNF40 (Cell Signaling, #12187, 1:1000), anti-HUWE1 (Cell Signaling, #5695, 1:1000) anti-TRIM33 (Cell Signaling, #13387, 1:1000), anti-STUB1 (Cell Signaling, #2080, 1:1000), anti-OCT4 (Abcam, #ab19857, 1:5000), anti-SOX2 (Abcam, #97959, 1:1000), anti-PAX6 (Stem Cell Technologies, #60094, 1:1000), anti-NESTIN (Stem Cell Technologies, # 60091, 1:1000), anti-PSMD3 (Proteintech, # 12054-1-AP, 1:1000), anti-EXOC5 (Proteintech, # 17593-1-AP, 1:1000) and anti-ß-actin (Abcam, #8226, 1:10000). Uncropped versions of western blots are presented in Supplementary Fig. 13.
Immunocytochemistry
Cells were fixed with paraformaldehyde (4% in PBS) for 20 min, followed by permeabilization (0.2% Triton X-100 in PBS for 10 min) and blocking (3% BSA in 0.2% Triton X-100 in PBS for 10 min). Human cells were incubated in primary antibody for 1,5 h at room temperature with anti-OCT4 (Stem Cell Technology, #60093, 1:200), anti-PAX6 (Stem Cell Technology, #60094, 1:300) or anti-SOX1 (Stem Cell Technology, #60095, 1:100). Then, cells were washed with PBS and incubated with secondary antibody (Alexa Fluor 488 goat anti-mouse (Thermo Fisher Scientific, A-11029), Alexa Fluor 568 F(ab’)2 Fragment of Goat Anti-Rabbit IgG (H + L) (Life Technologies, #A21069) and DAPI (Life Technologies, #1306) for 45 minutes at room temperature. PBS and distilled water wash were followed before the cover slips were mounted on Mowiol (Sigma, #324590).
RNA isolation and quantitative RT–PCR
Total RNA was isolated using RNAbee (Tel-Test Inc.). cDNA was generated from 1 μg of RNA using qScript Flex cDNA synthesis kit (Quantabio). SybrGreen real-time qPCR experiments were performed with a 1:20 dilution of cDNA using a CFC384 Real-Time System (Bio-Rad) following the manufacturer’s instructions. Data were analysed with the comparative 2ΔΔCt method using the geometric mean of ACTB and GAPDH as housekeeping genes. See Supplementary Information for details about the primers used for this assay.
26S proteasome fluorogenic peptidase assays
The in vitro assay of 26 S proteasome activities was performed as previously described136. Cells were collected in proteasome activity assay buffer (50 mM Tris-HCl, pH7.5, 250 mM sucrose, 5 mM MgCl2, 0.5 mM EDTA, 2 mM ATP and 1 mM DTT) and lysed by passing ten times through syringe needle (27 G). Then, lysate was centrifuged at 10,000 g for 10 min at 4 °C. 25 μg of total protein of cell lysates were transferred to a 96-well microtiter plate (BD Falcon) and incubated with the fluorogenic substrate Z-Gly-Gly-Leu-AMC (Enzo Lifescience). Fluorescence (380 nm excitation, 460 nm emission) was monitored on a microplate fluorometer (EnSpire, Perkin Elmer) every 5 min for 2 h at 37 °C.
Data availability
All the mass spectrometry proteomics data as well as the transcriptomic data are available in the Supplementary Information. All the other data are also available from the corresponding author upon request.
References
Jaenisch, R. & Young, R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell 132, 567–582 (2008).
Miura, T., Mattson, M. P. & Rao, M. S. Cellular lifespan and senescence signaling in embryonic stem cells. Aging Cell 3, 333–343 (2004).
Takahashi, K. et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 (2007).
Weinberger, L., Ayyash, M., Novershtern, N. & Hanna, J. H. Dynamic stem cell states: naive to primed pluripotency in rodents and humans. Nat Rev Mol Cell Biol 17, 155–169 (2016).
Lee, H. J., Gutierrez-Garcia, R. & Vilchez, D. Embryonic stem cells: a novel paradigm to study proteostasis? FEBS J 284, 391–398 (2017).
Buckley, S. M. et al. Regulation of pluripotency and cellular reprogramming by the ubiquitin-proteasome system. Cell Stem Cell 11, 783–798 (2012).
Garcia-Prat, L., Sousa-Victor, P. & Munoz-Canoves, P. Proteostatic and Metabolic Control of Stemness. Cell Stem Cell 20, 593–608 (2017).
Noormohammadi, A. et al. Mechanisms of protein homeostasis (proteostasis) maintain stem cell identity in mammalian pluripotent stem cells. Cell Mol Life Sci (2017).
Noormohammadi, A. et al. Somatic increase of CCT8 mimics proteostasis of human pluripotent stem cells and extends C. elegans lifespan. Nat Commun 7, 13649 (2016).
Schmidt, M. & Finley, D. Regulation of proteasome activity in health and disease. Biochim Biophys Acta 1843, 13–25 (2014).
Vilchez, D., Saez, I. & Dillin, A. The role of protein clearance mechanisms in organismal ageing and age-related diseases. Nat Commun 5, 5659 (2014).
Vilchez, D. et al. RPN-6 determines C. elegans longevity under proteotoxic stress conditions. Nature 489, 263–268 (2012).
Vilchez, D. et al. Increased proteasome activity in human embryonic stem cells is regulated by PSMD11. Nature 489, 304–308 (2012).
Pathare, G. R. et al. The proteasomal subunit Rpn6 is a molecular clamp holding the core and regulatory subcomplexes together. Proc Natl Acad Sci USA 109, 149–154 (2012).
Assou, S. et al. A gene expression signature shared by human mature oocytes and embryonic stem cells. BMC Genomics 10, 10 (2009).
Jang, J., Wang, Y., Kim, H. S., Lalli, M. A. & Kosik, K. S. Nrf2, a regulator of the proteasome, controls self-renewal and pluripotency in human embryonic stem cells. Stem Cells 32, 2616–2625 (2014).
Schroter, F. & Adjaye, J. The proteasome complex and the maintenance of pluripotency: sustain the fate by mopping up? Stem Cell Res Ther 5, 24 (2014).
Labbadia, J. & Morimoto, R. I. The biology of proteostasis in aging and disease. Annu Rev Biochem 84, 435–464 (2015).
Jeon, I. et al. Neuronal properties, in vivo effects, and pathology of a Huntington’s disease patient-derived induced pluripotent stem cells. Stem Cells 30, 2054–2062 (2012).
Hernebring, M., Brolen, G., Aguilaniu, H., Semb, H. & Nystrom, T. Elimination of damaged proteins during differentiation of embryonic stem cells. Proc Natl Acad Sci USA 103, 7700–7705 (2006).
Hernebring, M. et al. Removal of damaged proteins during ES cell fate specification requires the proteasome activator PA28. Sci Rep 3, 1381 (2013).
Okita, Y. & Nakayama, K. I. UPS delivers pluripotency. Cell Stem Cell 11, 728–730 (2012).
Tanaka, K. The proteasome: from basic mechanisms to emerging roles. Keio J Med 62, 1–12 (2013).
Balch, W. E., Morimoto, R. I., Dillin, A. & Kelly, J. W. Adapting proteostasis for disease intervention. Science 319, 916–919 (2008).
Powers, E. T., Morimoto, R. I., Dillin, A., Kelly, J. W. & Balch, W. E. Biological and chemical approaches to diseases of proteostasis deficiency. Annu Rev Biochem 78, 959–991 (2009).
Strikoudis, A., Guillamot, M. & Aifantis, I. Regulation of stem cell function by protein ubiquitylation. EMBO Rep 15, 365–382 (2014).
Suresh, B., Lee, J., Kim, K. S. & Ramakrishna, S. The Importance of Ubiquitination and Deubiquitination in Cellular Reprogramming. Stem Cells Int 2016, 6705927 (2016).
Vilchez, D., Simic, M. S. & Dillin, A. Proteostasis and aging of stem cells. Trends Cell Biol 24, 161–170 (2014).
Werner, A., Manford, A. G. & Rape, M. Ubiquitin-Dependent Regulation of Stem Cell Biology. Trends Cell Biol (2017).
Adams, J. The proteasome: structure, function, and role in the cell. Cancer Treat Rev 29(Suppl 1), 3–9 (2003).
Finley, D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem 78, 477–513 (2009).
Sussman, R. T. et al. The epigenetic modifier ubiquitin-specific protease 22 (USP22) regulates embryonic stem cell differentiation via transcriptional repression of sex-determining region Y-box 2 (SOX2). J Biol Chem 288, 24234–24246 (2013).
Zhang, X. Y. et al. The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Mol Cell 29, 102–111 (2008).
Fuchs, G. et al. RNF20 and USP44 regulate stem cell differentiation by modulating H2B monoubiquitylation. Mol Cell 46, 662–673 (2012).
Huang, Z. et al. Deubiquitylase HAUSP stabilizes REST and promotes maintenance of neural progenitor cells. Nat Cell Biol 13, 142–152 (2011).
Zhao, X. et al. The HECT-domain ubiquitin ligase Huwe1 controls neural differentiation and proliferation by destabilizing the N-Myc oncoprotein. Nat Cell Biol 10, 643–653 (2008).
You, K. T., Park, J. & Kim, V. N. Role of the small subunit processome in the maintenance of pluripotent stem cells. Genes Dev 29, 2004–2009 (2015).
Dupont, S. et al. FAM/USP9x, a deubiquitinating enzyme essential for TGFbeta signaling, controls Smad4 monoubiquitination. Cell 136, 123–135 (2009).
Metzger, M. B., Hristova, V. A. & Weissman, A. M. HECT and RING finger families of E3 ubiquitin ligases at a glance. J Cell Sci 125, 531–537 (2012).
de Bie, P. & Ciechanover, A. Ubiquitination of E3 ligases: self-regulation of the ubiquitin system via proteolytic and non-proteolytic mechanisms. Cell Death Differ 18, 1393–1402 (2011).
Tran, H. et al. HectD1 E3 ligase modifies adenomatous polyposis coli (APC) with polyubiquitin to promote the APC-axin interaction. J Biol Chem 288, 3753–3767 (2013).
Chuang, J. H., Tung, L. C. & Lin, Y. Neural differentiation from embryonic stem cells in vitro: An overview of the signaling pathways. World J Stem Cells 7, 437–447 (2015).
Fernandez, A. et al. The WNT receptor FZD7 is required for maintenance of the pluripotent state in human embryonic stem cells. Proc Natl Acad Sci USA 111, 1409–1414 (2014).
Patel, M. S. & Roche, T. E. Molecular biology and biochemistry of pyruvate dehydrogenase complexes. FASEB J 4, 3224–3233 (1990).
Zhang, J., Nuebel, E., Daley, G. Q., Koehler, C. M. & Teitell, M. A. Metabolic regulation in pluripotent stem cells during reprogramming and self-renewal. Cell Stem Cell 11, 589–595 (2012).
Kadamb, R., Mittal, S., Bansal, N., Batra, H. & Saluja, D. Sin3: insight into its transcription regulatory functions. Eur J Cell Biol 92, 237–246 (2013).
Saunders, A. et al. The SIN3A/HDAC Corepressor Complex Functionally Cooperates with NANOG to Promote Pluripotency. Cell Rep 18, 1713–1726 (2017).
Mallm, J. P. & Rippe, K. Aurora Kinase B Regulates Telomerase Activity via a Centromeric RNA in Stem Cells. Cell Rep 11, 1667–1678 (2015).
Johnson, M. T., Yang, H. S., Magnuson, T. & Patel, M. S. Targeted disruption of the murine dihydrolipoamide dehydrogenase gene (Dld) results in perigastrulation lethality. Proc Natl Acad Sci USA 94, 14512–14517 (1997).
Diao, Y. et al. Pax3/7BP is a Pax7- and Pax3-binding protein that regulates the proliferation of muscle precursor cells by an epigenetic mechanism. Cell Stem Cell 11, 231–241 (2012).
Guerrier, S. et al. The F-BAR domain of srGAP2 induces membrane protrusions required for neuronal migration and morphogenesis. Cell 138, 990–1004 (2009).
Beaulieu, C. L. et al. Intellectual disability associated with a homozygous missense mutation in THOC6. Orphanet J Rare Dis 8, 62 (2013).
Sanchez-Tena, S., Cubillos-Rojas, M., Schneider, T. & Rosa, J. L. Functional and pathological relevance of HERC family proteins: a decade later. Cell Mol Life Sci 73, 1955–1968 (2016).
Ji, Y. et al. The ancestral gene for transcribed, low-copy repeats in the Prader-Willi/Angelman region encodes a large protein implicated in protein trafficking, which is deficient in mice with neuromuscular and spermiogenic abnormalities. Hum Mol Genet 8, 533–542 (1999).
Arimitsu, N. et al. p125/Sec. 23-interacting protein (Sec. 23ip) is required for spermiogenesis. FEBS Lett 585, 2171–2176 (2011).
Jones, J. & Macdonald, P. M. Neurl4 contributes to germ cell formation and integrity in Drosophila. Biol Open 4, 937–946 (2015).
Al-Hakim, A. K., Bashkurov, M., Gingras, A. C., Durocher, D. & Pelletier, L. Interaction proteomics identify NEURL4 and the HECT E3 ligase HERC2 as novel modulators of centrosome architecture. Mol Cell Proteomics 11(M111), 014233 (2012).
Kuhnle, S. et al. Physical and functional interaction of the HECT ubiquitin-protein ligases E6AP and HERC2. J Biol Chem 286, 19410–19416 (2011).
Imai, Y. et al. The Parkinson’s Disease-Associated Protein Kinase LRRK2 Modulates Notch Signaling through the Endosomal Pathway. PLoS Genet 11, e1005503 (2015).
Emmott, E. & Goodfellow, I. Identification of protein interaction partners in mammalian cells using SILAC-immunoprecipitation quantitative proteomics. J Vis Exp (2014).
Charrier, C. et al. Inhibition of SRGAP2 function by its human-specific paralogs induces neoteny during spine maturation. Cell 149, 923–935 (2012).
Callaerts, P., Halder, G. & Gehring, W. J. PAX-6 in development and evolution. Annu Rev Neurosci 20, 483–532 (1997).
Lee, H. J. et al. A post-transcriptional program coordinated by CSDE1 prevents intrinsic neural differentiation of human embryonic stem cells. Nat Commun 8, 1456 (2017).
Jang, J. H. FIGC, a novel FGF-induced ubiquitin-protein ligase in gastric cancers. FEBS Lett 578, 21–25 (2004).
Ashton-Beaucage, D. et al. The Deubiquitinase USP47 Stabilizes MAPK by Counteracting the Function of the N-end Rule ligase POE/UBR4 in Drosophila. PLoS Biol 14, e1002539 (2016).
Hong, J. H. et al. KCMF1 (potassium channel modulatory factor 1) Links RAD6 to UBR4 (ubiquitin N-recognin domain-containing E3 ligase 4) and lysosome-mediated degradation. Mol Cell Proteomics 14, 674–685 (2015).
Jiang, J., Mohan, P. & Maxwell, C. A. The cytoskeletal protein RHAMM and ERK1/2 activity maintain the pluripotency of murine embryonic stem cells. PLoS One 8, e73548 (2013).
Luzzani, C. et al. Modulation of chromatin modifying factors’ gene expression in embryonic and induced pluripotent stem cells. Biochem Biophys Res Commun 410, 816–822 (2011).
Tasaki, T., Sriram, S. M., Park, K. S. & Kwon, Y. T. The N-end rule pathway. Annu Rev Biochem 81, 261–289 (2012).
Zimmerman, S. W. et al. Identification and characterization of RING-finger ubiquitin ligase UBR7 in mammalian spermatozoa. Cell Tissue Res 356, 261–278 (2014).
Zhong, J. F. et al. Gene regulation networks related to neural differentiation of hESC. Gene Expr 14, 23–34 (2007).
Yamane, M., Fujii, S., Ohtsuka, S. & Niwa, H. Zscan10 is dispensable for maintenance of pluripotency in mouse embryonic stem cells. Biochem Biophys Res Commun 468, 826–831 (2015).
Huibregtse, J. M., Scheffner, M. & Howley, P. M. Cloning and expression of the cDNA for E6-AP, a protein that mediates the interaction of the human papillomavirus E6 oncoprotein with p53. Mol Cell Biol 13, 775–784 (1993).
Williams, C. A. et al. Angelman syndrome. Curr Probl Pediatr 25, 216–231 (1995).
Buiting, K. Prader-Willi syndrome and Angelman syndrome. Am J Med Genet C Semin Med Genet 154C, 365–376 (2010).
Bolton, P. F. et al. The phenotypic manifestations of interstitial duplications of proximal 15q with special reference to the autistic spectrum disorders. Am J Med Genet 105, 675–685 (2001).
Khatri, N. et al. The autism protein Ube3A/E6AP remodels neuronal dendritic arborization via caspase-dependent microtubule destabilization. J Neurosci (2017).
Kuhnle, S., Mothes, B., Matentzoglu, K. & Scheffner, M. Role of the ubiquitin ligase E6AP/UBE3A in controlling levels of the synaptic protein Arc. Proc Natl Acad Sci USA 110, 8888–8893 (2013).
Miura, K. et al. Neurobehavioral and electroencephalographic abnormalities in Ube3a maternal-deficient mice. Neurobiol Dis 9, 149–159 (2002).
Mishra, A., Godavarthi, S. K. & Jana, N. R. UBE3A/E6-AP regulates cell proliferation by promoting proteasomal degradation of p27. Neurobiol Dis 36, 26–34 (2009).
Mishra, A. et al. E6-AP association promotes SOD1 aggresomes degradation and suppresses toxicity. Neurobiol Aging 34(1310), e1311–1323 (2013).
Mishra, A., Godavarthi, S. K., Maheshwari, M., Goswami, A. & Jana, N. R. The ubiquitin ligase E6-AP is induced and recruited to aggresomes in response to proteasome inhibition and may be involved in the ubiquitination of Hsp70-bound misfolded proteins. J Biol Chem 284, 10537–10545 (2009).
Toh, C. X. et al. RNAi Reveals Phase-Specific Global Regulators of Human Somatic Cell Reprogramming. Cell Rep 15, 2597–2607 (2016).
Huang, Y. Y., Lu, H., Liu, S., Droz-Rosario, R. & Shen, Z. Requirement of mouse BCCIP for neural development and progenitor proliferation. PLoS One 7, e30638 (2012).
Lim, H. J. et al. Regulation of c-Myc Expression by Ahnak Promotes Induced Pluripotent Stem Cell Generation. J Biol Chem 291, 752–761 (2016).
Schroer, T. A. Dynactin. Annu Rev Cell Dev Biol 20, 759–779 (2004).
Williams, S. E., Beronja, S., Pasolli, H. A. & Fuchs, E. Asymmetric cell divisions promote Notch-dependent epidermal differentiation. Nature 470, 353–358 (2011).
Inoue, D. et al. Dzip3 regulates developmental genes in mouse embryonic stem cells by reorganizing 3D chromatin conformation. Sci Rep 5, 16567 (2015).
Freund, A. et al. Proteostatic control of telomerase function through TRiC-mediated folding of TCAB1. Cell 159, 1389–1403 (2014).
Lopez, T., Dalton, K. & Frydman, J. The Mechanism and Function of Group II Chaperonins. J Mol Biol 427, 2919–2930 (2015).
Vander Heiden, M. G., Cantley, L. C. & Thompson, C. B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 (2009).
Varum, S. et al. Energy metabolism in human pluripotent stem cells and their differentiated counterparts. PLoS One 6, e20914 (2011).
Uchiki, T. et al. The ubiquitin-interacting motif protein, S5a, is ubiquitinated by all types of ubiquitin ligases by a mechanism different from typical substrate recognition. J Biol Chem 284, 12622–12632 (2009).
Kleijnen, M. F. et al. The hPLIC proteins may provide a link between the ubiquitination machinery and the proteasome. Mol Cell 6, 409–419 (2000).
Lee, S. Y. et al. Ube3a, the E3 ubiquitin ligase causing Angelman syndrome and linked to autism, regulates protein homeostasis through the proteasomal shuttle Rpn10. Cell Mol Life Sci 71, 2747–2758 (2014).
Tomaic, V. & Banks, L. Angelman syndrome-associated ubiquitin ligase UBE3A/E6AP mutants interfere with the proteolytic activity of the proteasome. Cell Death Dis 6, e1625 (2015).
Efroni, S. et al. Global transcription in pluripotent embryonic stem cells. Cell Stem Cell 2, 437–447 (2008).
Kim, Y. D. et al. The unique spliceosome signature of human pluripotent stem cells is mediated by SNRPA1, SNRPD1, and PNN. Stem Cell Res 22, 43–53 (2017).
Botti, V. et al. Cellular differentiation state modulates the mRNA export activity of SR proteins. J Cell Biol 216, 1993–2009 (2017).
Fang, X. et al. Landscape of the SOX2 protein-protein interactome. Proteomics 11, 921–934 (2011).
Tan, Y. et al. MFG-E8 Is Critical for Embryonic Stem Cell-Mediated T Cell Immunomodulation. Stem Cell Reports 5, 741–752 (2015).
Lee, S. H., Lee, Y. J., Park, S. W., Kim, H. S. & Han, H. J. Caveolin-1 and integrin beta1 regulate embryonic stem cell proliferation via p38 MAPK and FAK in high glucose. J Cell Physiol 226, 1850–1859 (2011).
Heck, J. W., Cheung, S. K. & Hampton, R. Y. Cytoplasmic protein quality control degradation mediated by parallel actions of the E3 ubiquitin ligases Ubr1 and San1. Proc Natl Acad Sci USA 107, 1106–1111 (2010).
Lee, J. T. & Gu, W. The multiple levels of regulation by p53 ubiquitination. Cell Death Differ 17, 86–92 (2010).
Rubenstein, E. M. & Hochstrasser, M. Redundancy and variation in the ubiquitin-mediated proteolytic targeting of a transcription factor. Cell Cycle 9, 4282–4285 (2010).
Xie, Y., Rubenstein, E. M., Matt, T. & Hochstrasser, M. SUMO-independent in vivo activity of a SUMO-targeted ubiquitin ligase toward a short-lived transcription factor. Genes Dev 24, 893–903 (2010).
Ingolia, N. T., Lareau, L. F. & Weissman, J. S. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147, 789–802 (2011).
Savic, N. et al. lncRNA maturation to initiate heterochromatin formation in the nucleolus is required for exit from pluripotency in ESCs. Cell Stem Cell 15, 720–734 (2014).
Jacinto, F. V., Benner, C. & Hetzer, M. W. The nucleoporin Nup153 regulates embryonic stem cell pluripotency through gene silencing. Genes Dev 29, 1224–1238 (2015).
Allen, N. D. & Baird, D. M. Telomere length maintenance in stem cell populations. Biochim Biophys Acta 1792, 324–328 (2009).
Zeng, S. et al. Telomerase-mediated telomere elongation from human blastocysts to embryonic stem cells. J Cell Sci 127, 752–762 (2014).
Cho, J. et al. LIN28A is a suppressor of ER-associated translation in embryonic stem cells. Cell 151, 765–777 (2012).
Peng, S. et al. Genome-wide studies reveal that Lin28 enhances the translation of genes important for growth and survival of human embryonic stem cells. Stem Cells 29, 496–504 (2011).
Wilbert, M. L. et al. LIN28 binds messenger RNAs at GGAGA motifs and regulates splicing factor abundance. Mol Cell 48, 195–206 (2012).
Xu, B., Zhang, K. & Huang, Y. Lin28 modulates cell growth and associates with a subset of cell cycle regulator mRNAs in mouse embryonic stem cells. RNA 15, 357–361 (2009).
Zhang, J. et al. LIN28 Regulates Stem Cell Metabolism and Conversion to Primed Pluripotency. Cell Stem Cell 19, 66–80 (2016).
Iconomou, M. & Saunders, D. N. Systematic approaches to identify E3 ligase substrates. Biochem J 473, 4083–4101 (2016).
O’Connor, H. F. & Huibregtse, J. M. Enzyme-substrate relationships in the ubiquitin system: approaches for identifying substrates of ubiquitin ligases. Cell Mol Life Sci 74, 3363–3375 (2017).
Sheng, Y. et al. Molecular recognition of p53 and MDM2 by USP7/HAUSP. Nat Struct Mol Biol 13, 285–291 (2006).
Pierce, N. W., Kleiger, G., Shan, S. O. & Deshaies, R. J. Detection of sequential polyubiquitylation on a millisecond timescale. Nature 462, 615–619 (2009).
Jain, A. K. & Barton, M. C. Regulation ofp53: TRIM24 enters the RING. Cell Cycle 8, 3668–3674 (2009).
Xu, P. et al. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell 137, 133–145 (2009).
Ding, W. X. et al. Linking of autophagy to ubiquitin-proteasome system is important for the regulation of endoplasmic reticulum stress and cell viability. Am J Pathol 171, 513–524 (2007).
Pandey, U. B. et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature 447, 859–863 (2007).
Chambers, S. M. et al. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol 27, 275–280 (2009).
Rao, J. et al. Stepwise Clearance of Repressive Roadblocks Drives Cardiac Induction in Human ESCs. Cell Stem Cell 18, 554–556 (2016).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Ran, F. A. et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154, 1380–1389 (2013).
Rappsilber, J., Ishihama, Y. & Mann, M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem 75, 663–670 (2003).
Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26, 1367–1372 (2008).
Cox, J. et al. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics 13, 2513–2526 (2014).
Cox, J. & Mann, M. 1D and 2D annotation enrichment: a statistical method integrating quantitative proteomics with complementary high-throughput data. BMC Bioinformatics 13(Suppl 16), S12 (2012).
Li, W. et al. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle’s dynamics and signaling. PLoS One 3, e1487 (2008).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Trapnell, C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7, 562–578 (2012).
Kisselev, A. F. & Goldberg, A. L. Monitoring activity and inhibition of 26S proteasomes with fluorogenic peptide substrates. Methods Enzymol 398, 364–378 (2005).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13, 2498–2504 (2003).
Mi, H., Muruganujan, A., Casagrande, J. T. & Thomas, P. D. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc 8, 1551–1566 (2013).
Acknowledgements
This work was supported by the European Research Council (ERC Starting Grant-677427 StemProteostasis), the Deutsche Forschungsgemeinschaft (DFG) (CECAD), and the Alexander von Humboldt Stiftung. We thank Christina Schindler for her technical support. We thank the CECAD Proteomics Facility and the Cologne Center for Genomics (CCG) for their advice and contribution in proteomics and RNA sequencing experiments, respectively.
Author information
Authors and Affiliations
Contributions
I.S. and S.K. performed all of the experiments, most of the data analysis and interpretation through discussions with D.V. R.G. performed the Cytoscape analysis. C.D. performed bioinformatic analysis of transcriptomics data and some proteomics data. D.V. planned and supervised the project. The manuscript was written by I.S. and D.V. All the authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Saez, I., Koyuncu, S., Gutierrez-Garcia, R. et al. Insights into the ubiquitin-proteasome system of human embryonic stem cells. Sci Rep 8, 4092 (2018). https://doi.org/10.1038/s41598-018-22384-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-22384-9
This article is cited by
-
HERC2 promotes inflammation-driven cancer stemness and immune evasion in hepatocellular carcinoma by activating STAT3 pathway
Journal of Experimental & Clinical Cancer Research (2023)
-
Effects of low doses of methylmercury (MeHg) exposure on definitive endoderm cell differentiation in human embryonic stem cells
Archives of Toxicology (2023)
-
Fbxw11 impairs the repopulation capacity of hematopoietic stem/progenitor cells
Stem Cell Research & Therapy (2022)
-
The E3 ubiquitin ligase HECTD1 contributes to cell proliferation through an effect on mitosis
Scientific Reports (2022)
-
Protein clearance strategies for disease intervention
Journal of Neural Transmission (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.