Abstract
Serratia marcescens is one of the important nosocomial pathogens which rely on quorum sensing (QS) to regulate the production of biofilm and several virulence factors. Hence, blocking of QS has become a promising approach to quench the virulence of S. marcescens. For the first time, QS inhibitory (QSI) and antibiofilm potential of Actinidia deliciosa have been explored against S. marcescens clinical isolate (CI). A. deliciosa pulp extract significantly inhibited the virulence and biofilm production without any deleterious effect on the growth. Vanillic acid was identified as an active lead responsible for the QSI activity. Addition of vanillic acid to the growth medium significantly affected the QS regulated production of biofilm and virulence factors in a concentration dependent mode in S. marcescens CI, ATCC 14756 and MG1. Furthermore vanillic acid increased the survival of Caenorhabditis elegans upon S. marcescens infection. Proteomic analysis and mass spectrometric identification of differentially expressed proteins revealed the ability of vanillic acid to modulate the expression of proteins involved in S-layers, histidine, flagellin and fatty acid production. QSI potential of the vanillic acid observed in the current study paves the way for exploring it as a potential therapeutic candidate to treat S. marcescens infections.
Similar content being viewed by others
Introduction
Serratia marcescens is an opportunistic human pathogen responsible for severe nosocomial infections1. Though S. marcescens was previously viewed as an innocuous organism, later on, it has been reported as an etiological agent of hospital-associated infections including septicemia, meningitis, endocarditis, urinary tract, respiratory tract, bloodstream and wound infections2. Prevalence of S. marcescens in healthcare settings is increasing parallel with the emergence of multi-drug resistance3,4. Biofilm formation is one of the primary underlying mechanisms responsible for antibiotic resistance in S. marcescens. This adherent nature of S. marcescens utilizes living as well as non-living surfaces such as biomedical devices to form the biofilm, finally leads to life-threatening infections5. A recent report states that biofilms produced by S. marcescens clinical isolates require supratherapeutic doses of kanamycin, gentamicin and chloramphenicol as minimum inhibitory concentration, which is 10, 100 and 1000 times higher concentration respectively, to kill the planktonic cells6.
Pathogenicity and biofilm formation of S. marcescens is directed by the cell density mediated gene expression mechanism called quorum sensing (QS). Acyl-homoserine lactones (AHLs) are the well-known autoinducers in LuxI/R QS system of Gram-negative bacteria7. Strains of S. marcescens have been reported to utilize a wide range of AHL molecules to regulate the transcription of target genes involved in biofilm formation, swimming and swarming motility, production of red coloured pigment prodigiosin, extracellular virulence enzymes (lipase, protease and nuclease), hemolysin and surfactants7. Briefly, the most common QS signal molecule used by the genus Serratia is N-hexanoyl-L-homoserine lactone (C6-HSL). The Serratia spp. strain ATCC 390068, S. marcescens MG19 and S. marcescens strain 12 utilizes N-butanoyl homoserine lactone (C4-HSL)10 as a QS signal molecule. In addition to C6-HSL, S. marcescens SS-1 also produces N-(3-oxohexanoyl) homoserine lactone (3-Oxo-C6-HSL), N-heptanoyl homoserine lactone (C7-HSL) and N-octanoyl homoserine lactone (C8-HSL) to regulate its QS dependent virulence factor production8. Hence, quenching its QS through the bioactive compounds from natural sources is a promising alternative strategy to combat infections caused by S. marcescens. For example, compounds such as O-methyl ellagic acid11 and alpha-bisabolol12, inhibiting QS in S. marcescens have been known to reduce the protease, lipase, prodigiosin and biofilm formation.
In recent years, awareness of the consumption of fruits for health promotion has increased. Actinidia deliciosa (Kiwifruit) has been classified as an excellent source of vitamin C, dietary fibre, vitamin E and potassium, based on the US FDA’s definition13. Kiwifruit has been known for its antioxidant, cardiovascular preventive and laxative activities14. Furthermore, consumption of kiwifruit selectively enhances the growth of intestinal bacteria such as Bifidobacterium and Lactobacillus15. Since the QSI potential of kiwifruit has never been explored and the presence of a wide range of secondary metabolites in kiwifruit has eventually led our focus to explore its QSI potential against S. marcescens.
Results and Discussion
Evaluation of QSI potential of A. deliciosa pulp extract (ADPE) against S. marcescens and identification of active principle
A number of studies have shown that bacteria, fungi and plants produce QSI compounds to protect themselves and compete with invading organisms. Due to their non-toxic and multifunctional nature, QSIs from edible sources are gaining attention in augmenting antimicrobial therapy16. S. marcescens strains from environmental and clinical origins were able to produce the intracellular prodigiosin pigment. Since prodigiosin production is under the direct regulation of the QS signalling mechanism17; the QSI potential of ADPE was initially assessed by prodigiosin assay. ADPE exhibited concentration-dependent prodigiosin inhibitory activity (Fig. 1a). At a higher concentration (20%) ADPE greatly reduced the prodigiosin production (85%) in S. marcescens when compared to control (Fig. 1a). The prodigiosin inhibitory activity of ADPE clearly suggests the presence of QS inhibitor(s).
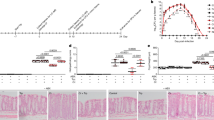
Quorum sensing inhibitory potential of ADPE against S. marcescens. The ADPE inhibited the QS regulated virulence factors such as biofilm formation, prodigiosin, protease, hemolysin and lipase (a) in a dose dependent manner without inhibiting the growth of S. marcescens (b). Light microscopic visualization of S. marcescens biofilm formed in the absence and presence of ADPE (c). Effect of solvent extracts of ADPEE against prodigiosin production and the growth (d) of S. marcescens. ADPEE-C exhibited the concentration dependent inhibition of biofilm formation, protease and lipase (e) without affecting the growth (f). Growth of S. marcescens was measured at 600 nm after 18 h incubation at 30 °C. Error bars represent standard deviations from the mean (n = 6 [biological triplicates in experimental duplicates]). Statistical significance was analyzed using one way ANOVA-Duncan’s post-hoc test. a, b, c and d indicate the significant difference p < 0.05, p < 0.01, p < 0.005 and p < 0.001, respectively.
In addition, dose-dependent inhibition of QS-regulated production of haemolysin (Fig. 1a) and biofilm formation (Fig. 1a) was observed with ADPE treatment without any reduction in the growth of marcescens (Fig. 1b). Light microscopic observation of biofilms formed in the presence and absence of ADPE further confirmed the potent antibiofilm activity (Fig. 1c). These results affirm the ability of ADPE to interfere with the QS mechanism of S. marcescens.
To identify the active lead(s) responsible for the QSI activity of ADPE, the ethanolic extract of ADPE (ADPEE) was extracted subsequently with hexane (ADPEE-H), chloroform (ADPEE-C), ethyl acetate (ADPEE-E) and butanol (ADPEE-B). The QSI activity of the dried solvent extracts was assessed based on its ability to reduce the QS-regulated prodigiosin production. The highest percentage of prodigiosin pigment reduction was observed in the ADPEE-C extract at 2 mg/mL (Fig. 1d). Henceforth, the effect of ADPEE-C extract on biofilm formation, production of protease and lipase production of S. marcescens was analysed. The ADPEE-C extract also showed a concentration-dependent inhibition of biofilm, protease and lipase (Fig. 1e) without any interference in the growth of S. marcescens (Fig. 1f).
The major constituents of ADPEE-C extract were identified using GC-MS analysis. The major components found in the ADPEE-C extract are vanillic acid (34.43%) and n-Hexadecanoic acid (11.85%) (Supplementary Fig. 1). Other major constituents present in the ADPEE- C are listed in supplementary Table 1. Vanillic acid is a well-known generally regarded as safe (GRAS) flavoring agent with antioxidant, anti-lipid peroxidative, anti-inflammatory and neuroprotective/cognitive effects.
Vanillic acid inhibits the QS regulated virulence factors and biofilm formation in S. marcescens
Pure vanillic acid was assayed for its QSI activity against S. marcescens CI, S. marcescens ATCC and S. marcescens MG1 virulence factors. Extracellular proteases of S. marcescens modulate the host’s immune response by inducing the inflammatory response18. Compounds/extracts inhibiting the production of protease can be used to potentiate the host’s innate immune response as well as antimicrobial therapy. In this context, the concentration-dependent decrease in the production of the secreted protease was observed in the cell free culture supernatant (CFCS) of S. marcescens CI (Fig. 2a), S. marcescens ATCC 14756 (Fig. 2b) and S. marcescens MG1 (Fig. 2c) grown in the presence of vanillic acid.
Quorum sensing inhibitory potential of active lead vanillic acid. Effect of pure vanillic acid on the QS regulated biofilm formation and prodigiosin, protease, hemolysin and lipase production of S. marcescens CI, S. marcescens ATCC and S. marcescens MG1 (a). Light (b) and CLSM [Three dimensional micrographs] (c) analyses corroborate the antibiofilm activity of vanillic acid. Effect of vanillic acid on the swarming motility of S. marcescens CI, S. marcescens ATCC and S. marcescens MG1 (d). Error bars represent standard deviations from the mean (n = 6 [biological triplicates in experimental duplicates]). Statistical significance was analyzed using one way ANOVA-Duncan’s post-hoc test. a, b, c and d indicate the significant difference p < 0.05, p < 0.01, p < 0.005 and p < 0.001, respectively.
Lipolytic enzymes are utilised by most of the bacterial pathogens to establish the infection by degrading the phospholipid bilayer of host cells and to manipulate the host cell signalling pathways19. Production of lipase was significantly reduced in all the three S. marcescens used in the present study. Briefly, 94 and 62% of inhibition of lipase production was noted in S. marcescens CI (Fig. 2a) and S. marcescens MG1 (Fig. 2b) respectively, at 250 µg/mL of vanillic acid. Whereas at 500 µg/mL of vanillic acid 65% of lipase inhibition was observed in S. marcescens ATCC 14756 (Fig. 2c). Haemolysin is a well-characterised virulence factor of S. marcescens. Vanillic acid reduced the haemolytic activity of S. marcescens CI and S. marcescens ATCC in a concentration-dependent mode. At 250 and 500 µg/mL of vanillic acid 68.75 and 56.52% of haemolysin production was observed in S. marcescens CI (Fig. 2a) and S. marcescens ATCC (Fig. 2b), respectively. These results show that vanillic acid has a potential to interfere with the QS mechanism of S. marcescens.
Biofilm formation has been considered as a major event in the establishment of chronic infections and confers extreme resistance to a broad spectrum of antibiotics. Vanillic acid has been reported for its ability to inhibit QS-dependent violacein pigment production in C. violaceum 20 and biofilm formation in Aeromonas hydrophila 21. Since QS is involved in the biofilm formation of S. marcescens 22, the anti-QS activity of vanillic acid is expected to have a significant negative effect on biofilm formation as well as swarming motility. In the present study, vanillic acid exhibited dose-dependent antibiofilm activity against S. marcescens CI (Fig. 2a), S. marcescens ATCC (Fig. 2b) and S. marcescens MG1 (Fig. 2c). Briefly, 63.6 and 57.64% of biofilm inhibition was noted at 250 µg/mL of vanillic acid in S. marcescens CI (Fig. 2a) and S. marcescens MG1 (Fig. 2c), respectively. At the same time, 50.84% (Fig. 2a) of biofilm inhibition was observed in S. marcescens ATCC grown in the presence of 500 µg/mL of vanillic acid.
In addition, biofilms of S. marcescens CI, S. marcescens ATCC and S. marcescens MG1 formed in the absence and presence of vanillic acid were observed under light microscope and the results revealed the reduction in biofilm formation and surface coverage (Fig. 2d). CLSM analysis also further confirmed the antibiofilm potential of vanillic acid (Fig. 2e). Swarming motility confers adaptive resistance to antibiotics and allows pathogenic bacteria to move across the moist, solid and viscous medium. Presence of vanillic acid in the growth medium was found to inhibit the swarming motility of all the three S. marcescens strains used in the present study (Fig. 2f).
As an ideal QSI is expected to have no/least interference with the basal growth of bacteria23, the effect of vanillic acid on the growth of S. marcescens CI, S. marcescens ATCC and S. marcescens MG1 was assessed by measuring the cell density at 600 nm. Results of growth measurement clearly suggest that vanillic acid does not have antibacterial activity at the tested concentrations. In addition, growth curve analysis was also carried out and the results revealed the non-antibacterial/bacteriostatic nature of vanillic acid at 125, 250 and 500 µg/mL (Fig. 3a,b and c).
Growth curve analysis depicting the non-antibacterial/bacteriostatic nature of S. marcescens CI (a), S. marcescens ATCC (b) and S. marcescens MG1 (c) grown in the presence and absence of vanillic acid. Error bars represent standard deviations from the mean (n = 3 [biological triplicates]). FTIR analysis of EPS extracted from control and vanillic acid treated S. marcescens CI (d). Panel e, f and g depict the variations in the polysaccharide, nucleic acid, proteins and fatty acid region of EPS, respectively.
FT-IR analysis showed the presence of polysaccharides, nucleic acid, proteins and fatty acids in extracted EPS of S. marcescens control and vanillic acid treated (Fig. 3c). The presence of peaks at 1,200–900 cm−1 indicates the C–OH stretching and C–O–C, C–O ring vibrations of carbohydrates24,25 peaks at 1,250–1,220 cm−1 corresponds to the phosphodiester, free phosphate, and monoester phosphate functional groups of phosphodiester, DNA/RNA backbone, phospholipids and phospho sugars26. Peaks at 1,540 and 1,650 cm−1 indicated the N–H bending, C–N stretching of Amide II and C = O stretching, C–N bending of Amide I proteins, respectively25 (Fig. 3c). In addition, a peak at 3,000–2,800 cm−1 corresponds to C–H vibrations of the functional groups of fatty acids24,25. The polysaccharide region (1,200–900 cm−1) of vanillic acid treated EPS was found to be increased slightly. However reduction in the absorbance of a peak at 1120 cm−1–1140 cm−1 which corresponds to the C-O-C stretching vibrations of carbohydrates was observed in vanillic acid treated EPS. Whereas the decrease in the IR absorbance at 1,250–1,220 cm−1(nucleic acid)27,28, 1,540 and 1,650 cm−1(Amide II and Amide I proteins)26, 3,000–2,800 cm−1 (fatty acid)25,29 was observed in vanillic acid treated EPS. Hence alterations in the biofilm architecture upon vanillic acid treatment could be attributed to the reduction of nucleic acid, amide I and II proteins and fatty acid content of EPS. When compared to the control, vanillic acid treated EPS showed a decrease in the absorbance of a broad peak at 3,800–3,100 cm−1 corresponding to –OH group (responsible for the hydration of EPS). Comparison of FT-IR spectra of control and treated EPS revealed a noticeable decrease in the IR absorbance of the nucleic acid, protein and fatty acid content and not in the carbohydrate content of the EPS (Fig. 3c). In addition to carbohydrate, other macromolecules such as nucleic acid, protein and fatty acid are also the main constituents required for the formation of EPS matrix. The decrease in the nucleic acid, protein and fatty acid content of EPS upon vanillic acid treatment could be one of the underlying mechanisms of its antibiofilm activity. Furthermore, the EPS inhibitory potential of vanillic acid is expected to decrease the survival of S. marcescens under in vivo conditions.
In vivo anti-QS and antibiofilm activity of vanillic acid
C. elegans has been used as one of the simple and successful in vivo preclinical models to study the bacterial pathogenesis and screening of bioactive compounds with antibacterial, anti-QS and antibiofilm activities etc30. Anti-QS and antibiofilm agents have been already reported to rescue/increase the survival of C. elegans during pathogenic infection31,32. Hence, in the present study the in vivo anti-QS and antibiofilm activity of vanillic acid against S. marcescens CI, S. marcescens ATCC and S. marcescens MG1 are evaluates using C. elegans. Worm liquid-killing assay was used to evaluate the ability of the vanillic acid to rescue the C. elegans from S. marcescens infection. No significant difference in the survival of worms fed with E. coli OP50 + vehicle control and E. coli OP50 + 500 µg/mL of vanillic acid confirms the non-toxic nature of vanillic acid (Fig. 4a). In addition, results of killing assay revealed that S. marcescens CI (Fig. 4a) is more pathogenic to C. elegans than S. marcescens ATCC (Fig. 4b) and S. marcescens MG1 (Fig. 4c). Briefly, the complete killing (100%) was observed at 78 h in S. marcescens CI infected C. elegans (Fig. 4a). Whereas complete killing of C. elegans was observed at 90 h and 102 h upon S. marcescens ATCC (Fig. 4b) and S. marcescens MG1 (Fig. 4c), respectively. Interestingly, vanillic acid increases the survival of C. elegans during S. marcescens CI, S. marcescens ATCC and S. marcescens MG1 infection by 51 (Fig. 4a), 58 (Fig. 4b) and 64% (Fig. 4c), respectively. Inhibition of in vivo colonization and prodigiosin production was further confirmed using light microscopic analysis. From the light micrographs, it is clear that in the presence of vanillic acid S. marcescens CI (Fig. 4d) and S. marcescens ATCC (Fig. 4e) failed to produce prodigiosin and reduced intestinal colonization in C. elegans. Briefly, pink coloration/colonization was observed in the intestinal region of C. elegans upon S. marcescens CI (Fig. 4d) and S. marcescens ATCC (Fig. 4e) infection. Whereas no such coloration was noted in C. elegans in the presence of vanillic acid + S. marcescens CI (Fig. 4d) and vanillic acid + S. marcescens ATCC (Fig. 4e). In addition, C. elegans infected with S. marcescens CI (Fig. 4d), S. marcescens ATCC (Fig. 4e) and S. marcescens MG1 (Fig. 4f) exhibited damaged pharynx and internal hatching. Conversely, C. elegans infected with vanillic acid + S. marcescens strains showed typical pharynx and well-organized eggs in their uterus region (Fig. 4d,e and f). For further confirmation of the ability of the vanillic acid to inhibit intestinal colonization of S. marcescens CFU assay was carried out. Results showed a significant decrease in intestinal colonization of S. marcescens strains in the presence of vanillic acid when compared to control (Fig. 4g,h and i). From these observations, it is apparent that vanillic acid increases the survival of C. elegans by targeting biofilm formation and QS in S. marcescens.
Survival graph showing the ability of vanillic acid treatment to rescue C. elegans from S. marcescens CI (a), S. marcescens ATCC (b) and S. marcescens MG1 (c) infection. Light micrographs depict the inhibition of prodigiosin pigment (d & e) and intestinal colonization (d,e & f) of S. marcescens upon vanillic acid treatment when compared to control. CFU assay results further confirm the inhibition of intestinal colonization of S. marcescens strains (g,h & i). Error bars represent standard deviations from the mean (n = 3 [biological triplicates in experimental duplicates]). Statistical significance was analyzed using one way ANOVA-Duncan’s post-hoc test. a and b indicate the significant difference p < 0.05 and p < 0.01, respectively.
Effect of vanillic acid treatment on the cellular proteome of S. marcescens
The results of the virulence assays prompted us to study the underlying molecular mechanism of QS inhibition of vanillic acid in S. marcescens. To get an overview, cellular proteins (each 30 µg) from the exponential phase cultures of S. marcescens CI grown in the presence and absence of vanillic acid (250 μg/mL) were separated in SDS-PAGE. CBB stained SDS-PAGE gel showed interesting protein profile variations in the cellular proteome of S. marcescens cultured in the presence of vanillic acid when compared to the control. Then, the cellular proteome of control and vanillic acid treated S. marcescens CI were analysed using 2DGE in biological triplicates (Supplementary Fig. 2). The protein spots present in the control and treated gels were detected and matched using Image Master Platinum 7 software (GE Healthcare). Based on the densitometric analysis, among the detected spots (579), 27 spots and 21 spots were found to be down-regulated and upregulated by more than 1.5 fold, respectively (Fig. 5). Based on statistical significance (ANOVA 0.05), 19 down-regulated and 14 upregulated protein spots were selected for protein identification. Protein spots with more than 2 fold differential expression were identified using nano-LC MS/MS analysis. Furthermore, 1.9 to 1.5 fold differentially expressed protein spots were identified using MALDI-TOF/TOF analysis. The list of differentially expressed proteins and their functions are presented in Table 1. Gene ontology analysis of differentially expressed proteins revealed their involvement in the biosynthesis of flagella, amino acids, prodigiosin, outer membrane, carbohydrates and lipids of S. marcescens (Fig. 6).
Representative gel pictures depicting the analysis of intracellular protein extract of S. marcescens CI grown in the absence and presence of 250 µg/mL of vanillic acid using 2-D gel electrophoresis. Each 450 µg of protein extract from control and treated cells were subjected to isoelectric focusing and resolved based on molecular weight in 10–15% gradient SDS-PAGE and protein spots were stained with MS compatible colloidal CBB. Up regulated and down regulated spots are encircled with green and red, respectively.
Vanillic acid treatment down regulated the expression of surface layer protein (slaA) in S. marcescens by 4.68 fold. Protein surface layer (S-layers) present in Gram-positive and Gram-negative bacteria has been known to involve in cell stabilization (mechanical, thermal and osmotic)33, compartmentalisation, protection from host immunological defences (phagocytosis)34, trapping of ions and immobilization of proteins35. SlaA of S. marcescens is partially similar to the Caulobacter crescentus paracrystalline S-layer protein and is recognized and transported into extracellular medium by the Lip system36. Role of slaA in biofilm formation of S. marcescens has not been identified yet. However, S-layer has been shown to have a role in adherence and maintaining cell surface hydrophobicity (CSH) of certain Gram-positive and Gram-negative bacteria37 and is one of the determinants required for the biofilm formation. Hence, down-regulation of slaA by vanillic acid could modulate the CSH in S. marcescens which is expected to affect the biofilm formation. Furthermore, vanillic acid also down-regulated (−2.11 fold) the expression of flagellin and upregulated (+2.57 fold) the expression of the long polar fimbrial chaperone (LpfB). In S. marcescens, flagellin is synthesized by the flhDC operon and polymerized into flagella, whereas LpfB is involved in the organization of cell wall and pili. Disruption of flhDC operon in S. marcescens CH−1 has been shown to be defective in swarming motility38. Flagella and swarming motility required for the initial attachment of biofilm formation39. Hence, down-regulation of flagellin by vanillic acid could be responsible for the reduction in biofilm formation. In addition, flagellar M-ring protein was also down-regulated upon vanillic acid treatment. Aminohydrolases are involved in nucleotide and amino acid metabolism40. Proteomic analysis revealed a 2 fold down-regulation of the expression of amidohydrolase of S. marcescens upon vanillic acid treatment.
Expression of probable phospholipid-binding protein (MlaC) was found to be upregulated upon vanillic acid treatment. MlaC is involved in bacterial intermembrane phospholipid trafficking41 and mlaC mutant of Escherichia coli has been shown to have increased phospholipids content in the outer membrane42. Hence upregulation of MlaC by vanillic acid could alter the phospholipid content of S. marcescens outer membrane. Urocanate hydratase (HutU) is involved in histidine metabolism found to down-regulated upon vanillic acid treatment by 2.70 fold when compared to control. Biosynthesis of purines and pyrimidines depends on the availability of histidine and DNA synthesis is linked to e-DNA release, which is also required for the biofilm formation43. In addition, HutU has been reported to have biofilm formation of Acinetobacter oleivorans 44 and Acinetobacter baumannii 43. It is likely that down-regulation of HutU by vanillic acid could also be responsible for the reduction of biofilm formation in S. marcescens.
UniProtKB shows that uncharacterized protein A0A0U6CCF3 is 90% identical in terms of sequence similarity to the biofilm development protein (bsmB [A0A0N1UW53]) of S. marcescens subsp. marcescens Db11. In S. marcescens, bsmB has already been shown to regulate by AHL mediated QS and plays a crucial role in biofilm development22. Recently phytol has been reported to inhibit the biofilm formation in S. marcescens by targeting the expression of bsmB45. Hence, we hypothesized that down-regulation of uncharacterized protein A0A0U6CCF3 in S. marcescens CI by vanillic acid could be responsible for the biofilm inhibition.
Enzymes such as beta-ketoacyl synthase (PigJ) and 4-hydroxy- 2, 2′- bipyrrole-5-methanol synthase (PigH) involved in prodigiosin biosynthesis of S. marcescens 15 were found to be down-regulated upon vanillic acid treatment. These results further validate the prodigiosin inhibitory potential of vanillic acid. Furthermore, 3-deoxymannooctulosonate-8-phosphatase (KdsC) which is known to involve in the biosynthesis of lipopolysaccharide and bacterial outer membrane synthesis46 was found to be down-regulated upon vanillic acid treatment. Since components of outer membrane and LPS are immunogenic and required for biofilm formation in Gram-negative bacterial pathogens46,47, down regulation of KdsC by vanillic acid could also be responsible for the reduction in the biofilm of S. marcescens.
Proteomic analysis of vanillic acid treated S. marcescens revealed the upregulation of multiple stress resistance protein (bhsA), which is predicted to be involved in stress response mechanism. BhsA of S. marcescens is 52.1% similar to putative outer membrane protein ycfR (bhsA) of E. coli in terms of the sequence. In E. coli, deletion ycfR (bhsA) has been shown to increase the biofilm, aggregation and CSH48. Upregulation of bhsA upon vanillic acid treatment could affect the biofilm formation in S. marcescens. In addition, vanillic acid treatment was found to upregulate the expression of 60 kDa chaperonin (groL) protein. GroL prevents the misfolding and helps in refolding/proper folding of proteins synthesized under stress condition49. Beta-ketoacyl-acyl carrier protein synthases are involved in fatty acid biosynthesis in most of the bacteria. Hence they have been identified as one of the promising targets for broad-spectrum antibacterial agents50. Down-regulation of beta-ketoacyl-acyl carrier protein synthase I, acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha and beta-ketoacyl-acyl-carrier-protein synthase I, 6-deoxyerythronolide B synthase suggested the ability of vanillic acid to interfere the fatty acid biosynthesis of S. marcescens. Furthermore, the expression of 3-oxoacyl-[acyl-carrier-protein] reductase (fabG) of S. marcescens was down regulated by vanillic acid. FabG has already been reported to regulate the virulence of Pseudomonas syringae pv. tabaci through AHL and fatty acid biosynthesis51. Whereas, in P. aeruginosa fabG determines the acyl chain length of 3-oxo-C12-HSL52. Role of fabG in QS and biofilm formation of S. marcescens has not been established yet. To get a preliminary information global sequence alignment analysis was carried out using EMBOSS Needle and the results revealed that the fabG of S. marcescens shares 64.8 and 78.1% identity and similarity, respectively to fabG of P. syringae pv. tabaci (A0A0W0PM71) and 63.2 and 75.3% identity and similarity, respectively to fabG (O54438) of P. aeruginosa. Hence it is hypothesized that down-regulation of fabG by vanillic acid could have a negative effect on QS-regulated virulence factors production in S. marcescens. In addition to inhibition of fatty acid biosynthesis pathway, amino acid (arginine, proline, cysteine and methionine) and butanoate metabolism have been predicted to inhibit the biofilm formation53. In the present study also down-regulation (1.5 fold) of cysteine synthase was noted upon vanillic acid treatment. Furthermore, down-regulation in the expression of aspartate ammonia lyase (aspA) was observed upon vanillic acid treatment. AspA has been reported to involve in the conversion of L-aspartate into fumarate and ammonia53. AspA mediated ammonia generation has already been reported to increase the intracellular polyamines synthesis, which in turn alters the membrane permeability, increases resistance to antibiotics and oxidative stress54. In contrast, the expression of butyryl-CoA dehydrogenase of S. marcescens was upregulated upon vanillic acid treatment. These results revealed the ability of vanillic acid to modulate the expression of proteins involved in the fatty acid and amino acid biosynthesis in S. marcescens CI.
Mutants of Serratia plymuthica RVH1 lacking splI (AHL synthase), has been shown to modulate the mixed-acid and butanediol fermentation. Briefly, mutation in splI gene resulted in enhanced and decreased acid and butanediol production respectively, in S. plymuthica 55. Interestingly in the present study vanillic acid treatment resulted in the upregulation of 2, 3-butanediol dehydrogenase (budC) of S. marcescens, which is under the control of QS. For further confirmation, Methyl Red Voges Proskauer (MR-VP) test was carried out in the present investigation and the results revealed that, vanillic acid treatment enhances the acid production and decreases the butanediol production. Though, 2, 3-butanediol dehydrogenase (budC) is upregulated upon vanillic acid treatment, the production of butanediol is reduced when compared to control (Fig. 7a and b). Butanediol production in S. plymuthica RVH1 and S. marcescens MG1 is regulated by AHL mediated QS and the growth of S. marcescens was not altered in the presence of vanillic acid. Hence reduction of butanediol production in S. marcescens upon vanillic acid treatment suggests its ability to modulate the AHL synthesis.
MR-VP test results depicting the effect of vanillic acid treatment on acid (a) and butanediol (b) production in S. marcescens CI. Effect of different concentration of vanillic acid treatment on the CSH (c) and the growth of S. marcescens CI in the presence of 0.5 and 1 M NaCl (d). Error bars represent standard deviations from the mean (n = 3). *(p < 0.05) indicates the statistical value of vanillic acid treated compared to control. Error bars represent standard deviations from the mean (n = 3 [biological triplicates in experimental duplicates]). Statistical significance was analyzed using one way ANOVA-Duncan’s post-hoc test. a, band c indicate the significant difference p < 0.05, p < 0.01 and p < 0.005, respectively.
In addition, vanillic acid treatment down regulated the expression of signal recognition particle receptor (FtsY) protein by 1.5 fold. FtsY is actively involved in the translocation/insertion of nascent membrane proteins into the cytoplasmic membrane56. Furthermore, molecular chaperone OsmY was found to be down regulated upon vanillic acid treatment in S. marcescens. EMBOSS Needle global sequence alignment analysis revealed that OsmY is 91.7% identical and 95.6% similar in terms of sequence with transport-associated protein (A8G9G9) of Serratia proteamaculans and which is shown to interact with the lipid A 1-diphosphate synthase (lpxT) and outer membrane protein assembly factor (BamA) involved in LPS biosynthesis and assembly of outer membrane proteins, respectively [interaction can be found in STRING database]57. Down-regulation of FtsY and OsmY suggests the possible involvement of vanillic acid to modulate the transport of membrane proteins.
Vanillic acid treatment was found to upregulate the expression of aldehyde dehydrogenase B (+4.74 fold) in S. marcescens. Recently, aromatic aldehyde dehydrogenases from Sphingobium sp. strain SYK-6 have been reported to catalyze the conversion of vanillin to syringaldehyde58. Hence, upregulation of aldehyde dehydrogenase B could be the possible mechanism responsible for the survival of S. marcescens in the presence of vanillic acid. Oxidoreductase has been reported to regulate the biofilm formation and virulence in Salmonella enterica serovar Typhimurium59 and Burkholderia cenocepacia 60. In the present study, vanillic acid treatment was found to down-regulate the expression of putative oxidoreductase by 2.7 fold. In addition, vanillic acid treatment was found to modulate the expression of ferrous iron transporter B (+5.77 fold), delta-aminolevulinic acid dehydratase (+2.40 fold) and several uncharacterized proteins (Table 1).
Role of slaA in biofilm formation and osmotic stress resistance in S. marcescens has not been studied yet. However, surface layer proteins of Lactobacillus acidophilus ATCC 4356 have been shown to protect the cells form NaCl induced osmotic stress61 and are responsible for adherence and cell surface hydrophobicity (CSH)37. To this end, CSH assay was carried out and the results revealed the concentration dependent reduction in CSH of S. marcescens upon vanillic acid treatment (Fig. 7c). Reduction in the CSH hinted the possible role of slaA in the CSH of S. marcescens. Furthermore, in the present study, we have also evaluated the effect of vanillic acid treatment on the osmotic stress resistance of S. marcescens and the results revealed that vanillic acid treated cells are unable to grow normally in the presence of 0.5 and 1 M NaCl when compared to control. These results suggest the ability of vanillic acid to modulate the osmotic stress resistance in S. marcescens by down regulating the expression of slaA protein. Further analyses are required to unveil the role of slaA in biofilm formation and virulence of S. marcescens.
Summary
In conclusion, vanillic acid has been identified as an active principle responsible for QSI and antibiofilm potential of kiwifruit (A. deliciosa) against S. marcescens. Virulence assays revealed the concentration dependent inhibitory activity over QS regulated biofilm, protease, prodigiosin and lipase production and swarming motility of S. marcescens CI and S. marcescens ATCC. In addition, vanillic acid also inhibited the protease and lipase production and biofilm formation in non-pigmented S. marcescens MG1. In addition, light and CLSM microscopic analyses further confirmed the antibiofilm activity of vanillic acid against S. marcescens CI, S. marcescens ATCC and S. marcescens MG1 used in this study. Furthermore, FTIR analysis of EPS extracted from control and vanillic acid treated S. marcescens CI showed the reduction in the nucleic acid, amide I and II proteins and fatty acid content. In vivo assays confirms the ability of vanillic acid to rescue C. elegans form the S. marcescens CI, S. marcescens ATCC and S. marcescens MG1 infection by inhibiting the QS regulated virulence and biofilm formation. Mass spectrometric identification of differentially expressed proteins and gene ontology analyses revealed the ability of vanillic acid modulates to the proteins involved in S-layers, flagellin, amino acid and fatty acid production in S. marcescens CI. The present study suggests that vanillic acid with its non-toxic nature and QSI potential can serve as a choice to potentiate the treatment strategy to overcome the QS- and biofilm-mediated infections caused by S. marcescens.
Materials and Methods
Ethical statement
Sheep blood was collected from the Karaikudi municipality modern slaughter house, Karaikudi and used to evaluate the effect of vanillic acid treatment on the hemolysin production in S. marcescens. Normally sheep blood is discarded in the butchery and hence no specific ethical permission was needed.
Collection of plant material and preparation of A. deliciosa pulp extract (ADPE)
A. deliciosa (Kiwifruit Zespri International Limited, New Zealand) was collected from the local market and the pulp was ground and centrifuged to collect the A. deliciosa pulp extract (ADPE). The ADPE was filter sterilized using 0.2 µm on the membrane filter (Millipore Corp., USA).
Bacterial strain and growth conditions
S. marcescens Clinical isolate used in this study is a clinical strain isolated from a urine sample, identified by 16S rRNA gene sequencing (GenBank Accession Number: FJ584421). In addition, S. marcescens ATCC 14756 and S. marcescens MG1 was also used to evaluate the anti-QS and antibiofilm activity of vanillic acid.
To determine the effect of ADPE on QS regulated extracellular virulence factors, LB medium (1% tryptone, 1% sodium chloride and 0.5% yeast extract) supplemented with and without (10–20% v/v, in increments of 5%) filter sterilized ADPE was inoculated with 1% (v/v) of the overnight culture of S. marcescens (OD600nm = 0.5) and incubated at 30 °C for 18 h at 120 rpm. After incubation, the cell density of control and treated S. marcescens CI was measured at 600 nm followed by centrifugation at 12,000 rpm for 15 min at 4 °C to collect the cell free culture supernatant (CFCS). CFCSs were stored at −20 °C for protease, lipase and hemolysin assay. The cell pellet was used for prodigiosin assay. Experiments are performed in biological triplicates in experimental duplicates.
Measurement of prodigiosin
For extraction of prodigiosin, cell pellets were resuspended in 1 ml acidified ethanol (4% (v/v) of 1 M Hydrochloric acid (HCl) in ethanol) and vortexed vigorously. Acidified ethanol containing solubilized prodigiosin and cell debris was subjected to centrifugation at 10,000 rpm for 10 min and the optical density of the supernatant was measured at 534 nm. The percentage of prodigiosin inhibition was calculated using the formula17:
\({\rm{ \% }}\,{\rm{o}}{\rm{f}}\,{\rm{i}}{\rm{n}}{\rm{h}}{\rm{i}}{\rm{b}}{\rm{i}}{\rm{t}}{\rm{i}}{\rm{o}}{\rm{n}}=({\rm{c}}{\rm{o}}{\rm{n}}{\rm{t}}{\rm{r}}{\rm{o}}{\rm{l}}\,{{\rm{O}}{\rm{D}}}_{534{\rm{n}}{\rm{m}}}-{\rm{t}}{\rm{e}}{\rm{s}}{\rm{t}}\,O{D}_{534{\rm{n}}{\rm{m}}}/\text{control}\,{{\rm{O}}{\rm{D}}}_{534{\rm{n}}{\rm{m}}})\times 100.\)
Total protease assay
The casein-degrading proteolytic activity was assessed by incubating each 100 µL reaction mix containing 2 mg/mL of azocaesin in 0.05 M Tris-hydrochloride, 0.5 mM CaCl2 (pH 7.5) with 100 µl of control and ADPE treated S. marcescens CFCS at 37 °C for 15 min. A 500 µl of 10% trichloroacetic acid was added to each reaction tube to terminate the reaction, followed by incubation at −20 °C for 10 min and the tubes were centrifuged at 12000 rpm for 20 min. Finally, the absorbance of the supernatant was measured at 400 nm using Multi-Mode Microplate Reader (SpectraMax M3, US)12.
Lipase assay
A 100 µl of S. marcescens culture supernatant (control and ADPE treated) was incubated with 900 µl of reaction mixture containing 1 volume 0.3% (w/v) p-nitro phenyl palmitate in isopropanol and 9 volumes of 50 mM Na2PO4 buffer (pH 8.0) containing 0.2% (w/v) sodium deoxycholate and 0.1% (w/v) gummi arabicum for 1 h at room temperature in dark. An equal volume of 1 M Na2CO3 was added to each tube to terminate the lipolytic activity and then centrifuged at 12,000 rpm for 10 min at room temperature. The absorbance of the supernatant was measured at 410 nm62.
Hemolysin assay
Hemolysin production was measured in accordance with the method described earlier18. Equal volume of 2% sheep red blood cells (RBCs) in phosphate buffered saline (pH 7.4) was mixed with control and ADPE treated S. marcescens CFCS at 37 °C for 2 h. After incubation, the reaction mixture was centrifuged at 8,000 rpm for 5 min and the absorbance of the supernatant was measured at 530 nm63.
Biofilm inhibition assay
Anti-biofilm potential of ADPE extract was determined using the method adopted by our group previously64. The biofilm of S. marcescens was allowed to grow in LB medium in the presence (10–20% v/v, in increments of 5%) and absence of ADPE in 24-well polystyrene plate at 30 °C for 24 h. After incubation, the wells were washed with distilled water to remove the planktonic cells. The sessile cells were stained with 0.4% crystal violet stain (CV) (w/v) for 5 minutes followed by washing off the unstained dye using distilled water. Finally, 1 mL of absolute ethanol was added to each well to dissolve the CV present in the biofilm cells. The absorbance (OD570nm) was determined and percentage of biofilm inhibition was calculated using the formula:
Successive extraction of A. deliciosa pulp extract
Freeze-dried A. deliciosa pulp was ground into fine powder using a blender and extracted with 70% ethanol for 3 h at boiling point and filtered through Whatman filter paper No. 2. The filtered A. deliciosa pulp ethanolic extract (ADPEE) was evaporated under vacuum at 55 °C for the complete removal of water and ethanol. Then the ADPEE was dissolved in distilled water and extracted successively with hexane (ADPEE-H), chloroform (ADPEE-C), ethyl acetate (ADPEE-E), and n-butanol (ADPEE-B), finally leaving an aqueous fraction of ethanol extract (ACPEE-W)65. Above mentioned solvent extracts were dried under vacuum and assayed for their QSI activity against S. marcescens as mentioned previously.
GC-MS analysis of ADPEE-C
ADPEE-C extract exhibited concentration dependent inhibition of QS regulated virulence factors of S. marcescens. Thus ADPEE-C was analysed by SHIMADZU GCMS-138 QP2010 plus, containing Rxi®-5ms gas chromatograph column (ID 0.25 mm thickness 0.25 139 µm), coupled to a mass spectrometer mass detector (with 5% diphenyl and 95% 140 dimethylpolysiloxane) using standard GC-MS parameters. The constituents of ADPEE-C were identified by processing of the raw GC-MS data with the NIST software (National Institute of Standards and Technology, Gaithersburg, USA).
Effect of pure vanillic acid on the QS regulated virulence factors production of S. marcescens strains
Pure vanillic acid (Sigma, USA) was dissolved in ethanol (100 mg/mL). S. marcescens CI, S. marcescens ATCC and S. marcescens MG1 were grown in the presence and absence of 62.5, 125, 250 and 500 µg/mL vanillic acid and incubated at 30 °C for 18 h at 120 rpm. After incubation growth, prodigiosin, lipase, protease, hemolysin and biofilm formation were measured as mentioned previously.
CLSM analysis of biofilm
For CLSM analysis, biofilm slides were prepared by growing S. marcescens CI, S. marcescens ATCC and S. marcescens MG1 in the absence and the presence of 250 and 500 µg/mL of vanillic acid for 24 h at 30 °C. The biofilms formed on the glass slides were stained with 0.1% acridine orange for 1 min, washed with distilled water, air dried and visualized under CLSM (Carl Zeiss, Germany) at 20× magnification. CLSM images (N = 3) were obtained from the triplicate of control and treated biofilm and processed using Zen 2009 image software64.
Swarming motility assay
The effect of vanillic acid on the swarming motility was determined by placing 2 µL of overnight cultures of S. marcescens in the centre of swarm agar plates (peptone (5 g/L), glycerol (1% (v/v)) and agar (0.75%)). The plates were incubated at 25 °C for 20 h. The activity was determined by the decrease in the swarmed area radius5.
Extraction of EPS and FTIR analysis
EPS extraction was carried out in accordance with the previously published methods66. Briefly, each 100 mL of S. marcescens CI control and vanillic acid treated cultures were centrifuged at 7, 000 rpm for 15 min at 4 °C to collect the cells. Cell free culture supernatants were stored at −20 °C for cell free EPS extraction. The collected cell pellets were washed with wash buffer (10 mM Tris/HCl pH 8.0, 10 mM EDTA) and resuspended in 100 mL of isotonic extraction buffer (10 mM Tris/HCl pH 8.0, 10 mM EDTA, 0.5 mM NaCl) and incubated at 4 °C for 12 h. After incubation, isotonic buffer containing cells were vortexed for 5 min and centrifuged at 5000 rpm for 15 min to collect the supernatant containing cell bound EPS. Then, both CFCS containing cell free EPS and isotonic buffer containing cell bound EPS were pooled and mixed with 3 volumes of ice-cold ethanol and kept at −20 °C for 18 h for the precipitation of EPS. Precipitated EPS was collected by centrifugation at 12, 000 rpm for 30 min at 4 °C and dried under vacuum62. For FTIR analysis, each 2 mg of EPS from control and treated were mixed with 100 mg of potassium bromide (KBr) to prepare KBr-EPS pellet and infrared spectra were collected in the range of 400 to 4000 cm−1 using FTIR spectrometer (Bruker Tensor 27) and values were plotted as intensity versus wave number.
Caenorhabditis elegans maintenance
C. elegans wild type N2 worms were obtained from Caenorhabditis Genetic Centre (CGC) Minnesota, USA and maintained in Nematode Growth Medium (NGM) containing E. coli OP50 as a food source. Synchronised L4 stage animals were used for all the experimental. To obtain synchronised L4 stage worms, adult stage animals were bleached with 1:1 ratio of commercial bleach and 1 M KOH67.
Toxicity and liquid killing assay
To check the toxicity of vanillin, 15 numbers of synchronised L4 stage C. elegans were transferred to M9 buffer (500 µL) containing E. coli OP50 (~1000 CFU/mL), presence and absence of vanillin 250 µg/mL. Ethanol was used as a vehicle in all the experiments. To evaluate the anti-QS and antibiofilm activity of vanillic acid, 15 numbers of synchronised L4 stage C. elegans were transferred to 24 well microtitre plate containing 400 µl of M9 buffer + 100 µL of S. marcescens CI and S. marcescens MG1 culture with 0.3 OD at 600 nm and supplemented with and without vanillic acid 250 µg/mL. In the case of S. marcescens ATCC 14756, 500 µg/mL of vanillic acid was used. The assay was monitored continuously for every 6hrs interval until the complete death of C. elegans in control. The animals did not show any response to touch by the external stimuli like platinum worm picking loop then it was considered as dead67.
Microscopic observation
For microscopic observation, C. elegans were exposed to S. marcescens CI, S. marcescens ATCC and S. marcescens MG1, with and without vanillic acid for 24hrs. Then control and treated C. elegans were washed with M9 buffer thoroughly and placed on a microscopic slide with 1 mM Sodium azide and visualized under bright field microscope68.
Colony forming unit assay (CFU)
To access the intestinal colonization of S. marcescens CI, S. marcescens ATCC and S. marcescens MG1 in C. elegans CFU was performed. Briefly synchronised L4 stage C. elegans were exposed to S. marcescens CI, S. marcescens ATCC and S. marcescens MG1, with and without vanillin 250 µg/mL for 24hrs. After 24hrs, 20 numbers of exposed worms were washed thoroughly with M9 buffer containing 1 mM sodium azide to reduce the discharge of bacteria from the worm intestine and finally crushed/ground with 400 mg of 1.0 mm of silicon carbide and vortexed for 20 minutes. The final suspension was serially diluted and plated onto specific medium68.
Extraction of cellular proteins
For extraction of intracellular proteins, S. marcescens was grown in the absence and presence of 250 µg/mL of vanillic acid for 18 h at 30 °C with 120 rpm in conical flasks in biological triplicates and cells were harvested by centrifugation at 8000 rpm for 12 min at 4 °C and the resulting pellets were washed twice with 50 mM tris-HCl (pH 8.0). Cell pellets were resuspended in 50 mM tris-HCl (pH 8.0) supplemented with 1% of protease inhibitor cocktail and sonicated using Ultra-sonicator (Sonics VCX 750, USA) with the parameters of 35 KHz, 750 W and 35% amplitude for 5 min with pulse time of 10 sec (for both on and off cycles) followed by centrifugation at 13,000 rpm for 30 min at 4 °C to remove the cell debris. Supernatants containing cellular proteins were purified using phenol extraction and precipitated by acetone. Precipitated proteins were washed thrice with ice-cold acetone to wash off the remaining phenol residues and air dried. Finally, the air-dried protein pellets were dissolved in urea thiourea sample buffer (7 M urea, 2 M thiourea, 4% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate [CHAPS], 15 mM dithiothreitol [DTT] and 0.5% of immobilised pH gradient [IPG] buffer [pH 3–10]) and centrifuged at 13,000 rpm for 20 min at 4 °C and the concentration of proteins present in the samples were quantified by Bradford assay kit (BioRad). Each 450 µg of cellular proteins form control and vanillic acid treated S. marcescens were used for isoelectric focusing (IEF) in biological triplicates69.
Two-dimensional gel electrophoresis (2DGE) and image analysis
Protein samples (each 400 µg) from control and vanillic acid treated S. marcescens were diluted with urea thiourea rehydration buffer (7 M urea, 2 M thiourea and 2% [CHAPS]) supplemented with 12.5 mg/mL destreak reagent and 0.5% IPG buffer [pH 3–10] to a final volume of 350 µl. Immobiline DryStrip gel strips (18 cm, non-linear, pH 3–10) were rehydrated with thiourea rehydration buffer containing cellular proteins for 12 h at 20 °C and the rehydrated strips were iso-electrically focused in IPGphor 3 system using standard parameters. After IEF, IPG strips were subjected to reduction and alkylation with DTT and iodoacetamide (IAA), respectively. Then the strips were placed on 25 cm × 22 cm × 1 mm 10–15% gradient sodium dodecyl sulphate-polyacrylamide gels (SDS-PAGE), sealed with 0.3% agarose solution containing a trace amount of tracking dye Bromophenol blue and electrophoresis was carried out at 100 V for 1 h and 150 V until the dye front had reached the bottom of the SDS-PAGE gel in Ettan DALT six apparatus (GE Healthcare). After electrophoresis, the gels incubated in fixative solution (40% methanol, 10% glacial acetic acid (GAA) and 50% MilliQ H2O) for 3 h. Protein spots were visualized using mass spectrometry compatible colloidal coomassie brilliant blue (CBB) G-250 staining for 12 h and destained with MilliQ H2O until the background becomes clear. Then, image acquisition was done with Image Scanner III (GE Healthcare) and analysed by ImageMaster 2D Platinum 7.0 software (GE Healthcare)69.
Protein spots excision and in-gel trypsin digestion
The protein spots with 1.5 fold differential regulation (up/down) were excised from the gels and the excised gel plugs were destained with 50% acetonitrile (ACN) containing 25 mM ammonium bicarbonate (NH4HCO3) until the complete removal of CBB. Destained gel plugs were dehydrated in ACN, vacuum dried and rehydrated with 5 μL of digestion buffer (10 mM NH4HCO3 in 10% ACN) containing 400 ng of sequencing grade trypsin (Sigma Aldrich) on ice for 30 min. After rehydration with digestion buffer, gel plugs were flooded with 25 μL of overlay solution (40 mM NH4HCO3 in 10% ACN) and incubated at 37 °C for 16 h. After trypsin digestion, peptides were extracted with peptide extraction solution (0.1% trifluoroacetic acid (TFA) in 60% ACN) by sonication followed by complete dehydration of gel plugs with ACN. Collected peptides were pooled and vacuum dried for 90 min and stored at 4 °C70. For nano LC-MS/MS and MALDI-TOF/TOF analyses, vacuum dried peptides were purified using C18 zip tips (Merck Millipore) as per the manufacturer’s instructions.
Identification of differentially expressed proteins by nano LC-MS/MS and MALDI-TOF/TOF analyses
Protein spots with ≥2 fold differential regulation were analysed using nano reverse phase-LC (Thermo Scientific, USA) coupled with an Orbitrap Elite Mass spectrometer (Thermo Scientific, USA) using standard parameters71. The MS data acquisition was done in positive ion mode (m/z 350–4000 Da) using Xcalibur software (Version 2.2.SP1.48, Thermo Scientific, USA). Protein identification was done with Proteome Discoverer software v.1.4 (Thermo Scientific) using the following parameters: 2 missed cleavages, 10 ppm and 0.5 Da as precursor mass tolerance and fragment mass tolerance, respectively. In addition, cystine carbamidomethylation as fixed modification, methionine oxidation, N-terminal acetylation and phosphorylation (S, T, Y) as variable modification.
Protein spots with 1.5 to 1.9 fold differential regulation were analysed using MALDI-TOF/TOF analysis. Prior to analysis, internal calibration was carried out using TOF-Mix™ (LaserBio Labs, France). For MALDI-TOF/TOF analysis, 1 µL of purified peptides were mixed with 1 µL of the alpha-cyano-4-hydroxy cinnamic acid matrix (10 mg/mL) on MALDI target plate. Mass spectra were acquired in positive reflectron mode and the monoisotopic peak list (m/z range of 700–4000 Da) was used for protein identification using MS-Fit (http://prospector.ucsf.edu) online software using standard parameters69.
MR-VP test
S. marcescens was allowed to grow in MR-VP broth supplemented with and without 125 and 250 µg/mL of vanillic acid for 24 h. After incubation, to detect the acid production, methyl red indicator dye was added and the changes in the colour was visually observed and photographed. To detect the butanediol formation Barritt’s reagent A and B were added to the culture, mixed well and formation of red coloration was visually observed and photographed.
Osmotic sensitivity assay
To analyse the effect of vanillic acid treatment on the osmotic sensitivity, S. marcescens was allowed to grow in LB medium supplemented with 0.5 and 1 M NaCl in the presence and absence of 125 and 250 µg/mL of vanillic acid at 30 °C and cell density was measured periodically for 24 h with 2 h interval61.
Statistical analysis
All the experiments were performed independently in triplicate to confirm reproducibility. Data were analyzed by one-way analysis of variance (ANOVA) followed by Duncan’s post-hoc test with a significant P value of <0.05 using the SPSS (Chicago, IL, USA) statistical software package.
References
Gupta, N. et al. Outbreak of Serratia marcescens bloodstream Infections in patients receiving parenteral nutrition prepared by a compounding pharmacy. Clin. Infect. Dis. 59, 1–8, https://doi.org/10.1093/cid/ciu218 (2014).
Hejazi, A. & Falkiner, F. R. Serratia marcescens. J. Med. Microbiol. 46, 903–912, https://doi.org/10.1099/00222615-46-11-903 (1997).
Faidah, H. S., Ashgar, S. S., Barhameen, A. A., El-Said, H. M. & Elsawy, A. Serratia marcescens as opportunistic pathogen and the importance of continuous monitoring of nosocomial infection in Makah city, Saudi Arabia. Open J. Med. Microbiol. 5, 107–112, https://doi.org/10.4236/ojmm.2015.53013 (2015).
Gruber, T. M. et al. Pathogenicity of pan-drug-resistant Serratia marcescens harbouring blaNDM-1. J. Antimicrob. Chemother. 70, 1026–1030, https://doi.org/10.1093/jac/dku482 (2014).
Shanks, R. M. et al. A Serratia marcescens OxyR homolog mediates surface attachment and biofilm formation. J. Bacteriol. 189, 7262–7272, https://doi.org/10.1128/JB.00859-07 (2007).
Ray, C., Shenoy, A. T., Orihuela, C. J. & González-Juarbe, N. Killing of Serratia marcescens biofilms with chloramphenicol. Ann. Clin. Microbiol. Antimicrob. 16, 19, https://doi.org/10.1186/s12941-017-0192-2 (2017).
Van Houdt, R., Givskov, M. & Michiels, C. W. Quorum sensing in Serratia. FEMS. Microbiol. Rev. 31, 407–424, https://doi.org/10.1111/j.1574-6976.2007.00071.x (2007).
Horng, Y. T. et al. The LuxR family protein SpnR functions as a negative regulator of N-acylhomoserine lactone dependent quorum sensing in Serratia marcescens. Mol. Microbiol. 45, 1655–1671, https://doi.org/10.1046/j.1365-2958.2002.03117.x (2002).
Eberl, L. G. et al. Involvement of N-acyl-l-homoserine lactone autoinducers in controlling the multicellular behaviour of Serratia liquefaciens. Mol. Microbiol. 20, 127–136, https://doi.org/10.1111/j.1365-2958.1996.tb02495.x (1996).
Coulthurst, S. J., Williamson, N. R., Harris, A. K., Spring, D. R. & Salmond, G. P. Metabolic and regulatory engineering of Serratia marcescens: mimicking phage mediated horizontal acquisition of antibiotic biosynthesis and quorum-sensing capacities. Microbiology 152, 1899–1911, https://doi.org/10.1099/mic.0.28803-0 (2006).
Salini, R. & Pandian, S. K. Interference of quorum sensing in urinary pathogen Serratia marcescens by Anethum graveolens. Pathog. Dis. 73, ftv038, https://doi.org/10.1093/femspd/ftv038 (2015).
Sethupathy, S., Shanmuganathan, B., Devi, K. P. & Pandian, S. K. Alpha-bisabolol from brown macroalga Padina gymnospora mitigates biofilm formation and quorum sensing controlled virulence factor production in Serratia marcescens. J. Appl. Phycol. 28, 1987–1996, https://doi.org/10.1007/s10811-015-0717-z (2016).
Nishiyama, I. et al. Varietal difference in vitamin C content in the fruit of kiwifruit and other Actinidia species. J. Agric. Food Chem. 52, 5472–5475, https://doi.org/10.1021/jf049398z (2004).
Jung, K. A. et al. Cardiovascular protective properties of kiwifruit extracts in vitro. Biol. Pharm. Bull. 28, 1782–1785, https://doi.org/10.1248/bpb.28.1782 (2005).
Lee, Y. K., Low, K. Y., Siah, K., Drummond, L. M. & Gwee, K. A. Kiwifruit (Actinidia deliciosa) changes intestinal microbial profile. Microb. Ecol. Health. Dis. 23, eCollection, https://doi.org/10.3402/mehd.v23i0.18572 (2012).
Givskov, M. Beyond nutrition: health-promoting foods by quorum-sensing inhibition. Future. Microbiol. 7, 1025–1028, https://doi.org/10.2217/fmb.12.84 (2012).
Slater, H., Crow, M., Everson, L. & Salmond, G. P. Phosphate availability regulates biosynthesis of two antibiotics, prodigiosin and carbapenem, in Serratia via both quorum-sensing-dependent and -independent pathways. Mol. Microbiol. 47, 303–320, https://doi.org/10.1046/j.1365-2958.2003.03295.x (2003).
Kurz, C. L. et al. Virulence factors of the human opportunistic pathogen Serratia marcescens identified by in vivo screening. EMBO. J. 22, 1451–1460, https://doi.org/10.1093/emboj/cdg159 (2003).
Bender. J. & Flieger. A. Lipases as pathogenicity factors of bacterial pathogens of humans. Handbook of Hydrocarbon and Lipid Microbiology (ed. Timmis, K.N.) 3241–3258 (Springer Berlin Heidelberg, 2010).
Choo, J., Rukayadi, H. Y. & Hwang, J. K. Inhibition of bacterial quorum sensing by vanilla extract. Lett. Appl. Microbiol. 42, 637–641, https://doi.org/10.1111/j.1472-765X.2006.01928.x (2006).
Kappachery, S., Paul, D., Yoon, J. & Kweon, J. H. Vanillin, a potential agent to prevent biofouling of reverse osmosis membrane. Biofouling 26, 667–672, https://doi.org/10.1080/08927014.2010.506573 (2010).
Labbate, M. et al. Quorum sensing-controlled biofilm development in Serratia liquefaciens MG1. J. Bacteriol. 186, 692–698, https://doi.org/10.1128/JB.186.3.692-698.2004 (2010).
Rasmussen, T. B. & Givskov., M. Quorum sensing inhibitors: a bargain of effects. Microbiology 152, 895–904, https://doi.org/10.1099/mic.0.28601-0 (2006).
Helm, D., Labischinsky, H., Schallehn, G. & Naumann, D. Classification and identification of bacteria by Fourier-transform infrared spectroscopy. J. Gen. Microbiol. 137, 69–79, https://doi.org/10.1099/00221287-137-1-69 (1991).
Beech, I., Hanjagsit, L., Kalaji, M., Neal, A. L. & Zinkevich, V. Chemical and structural characterization of exopolymers produced by Pseudomonas sp. NCIMB 2021 in continuous culture. Microbiology 145, 1491–1497, https://doi.org/10.1099/13500872-145-6-1491 (1999).
Naumann D (2000) Infrared spectroscopy in microbiology. Wiley, Chichester.
Zeroual, W. et al. Monitoring of bacterial growth and structural analysis as probed by FT-IR spectroscopy. Biochim. Biophys. Acta. 1222, 171–178, https://doi.org/10.1016/0167-4889(94)90166-X (1994).
Schmitt., J., Nivens, D., White, D. C. & Flemming, H. C. Changes of biofilm properties in response to sorbed substances- An FT-IR/ ATR study. Water Sci. Technol. 32, S149–S155, https://doi.org/10.1016/0273-1223(96)00019-4 (1995).
Moreno, J. et al. Chemical and rheological properties of an extracellular polysaccharide produced by the cyanobacterium Anabaena sp. ATCC 33047. Biotechnol. Bioeng. 67, 283–290, https://doi.org/10.1002/(SICI)1097-0290(20000205)67:3<283::AID-BIT4>3.0.CO;2-H (2000).
Kim, W., Hendricks, G. L., Lee, K. & Mylonakis, E. An update on the use of C. elegans for preclinical drug discovery: screening and identifying anti-infective drugs. Expert Opin. Drug Disco. 12, 625–633, https://doi.org/10.1080/17460441.2017.1319358 (2017).
Viszwapriya, D., Subramenium, G. A., Prithika, U., Balamurugan, K. & Pandian, S. K. Betulin inhibits virulence and biofilm of Streptococcus pyogenes by suppressing ropB core regulon, sagA and dltA. Pathog. Dis. 74(pii), ftw088, https://doi.org/10.1093/femspd/ftw088 (2016).
Gowrishankar, S., Kamaladevi, A., Ayyanar, K. S., Balamurugan, K. & Pandian, S. K. Bacillus amyloliquefaciens-secreted cyclic dipeptide- cyclo (L-Leucyl- L-Prolyl) inhibits biofilm and virulence in methicillin-resistant Staphylococcus aureus. RSC Advances 5, 95788–95804, https://doi.org/10.1039/C5RA11641D (2015).
Engelhardt, H. Mechanism of osmoprotection by archaeal S-layers: a theoretical study. J. Struct. Biol. 160, 190–199, https://doi.org/10.1016/j.jsb.2007.08.004 (2007a).
Blaser, M. J. Role of the S-layer proteins of Campylobacter fetus in serum-resistance and antigenic variation: a model of bacterial pathogenesis. Am. J. Med. Sci. 306, 325–329 (1993).
Engelhardt, H. Are S-layers exoskeletons? The basic function of protein surface layers revisited. J. Struct. Biol. 160, 115–124, https://doi.org/10.1016/j.jsb.2007.08.003 (2007b).
Kawai, E., Akatsuka, H., Idei, A., Shibatani, T. & Omori, K. Serratia marcescens S‐layer protein is secreted extracellularly via an ATP‐binding cassette exporter, the Lip system. Mol. Microbiol. 27, 941–952, https://doi.org/10.1046/j.1365-2958.1998.00739.x (1998).
Van der Mei, H. C., Van de Belt-Gritter, B., Pouwels, P. H., Martinez, B. & Busscher, H. J. Cell surface hydrophobicity is conveyed by S-layer proteins—a study in recombinant lactobacilli. Colloids Surf. B Biointerfaces 28, 127–134, https://doi.org/10.1016/S0927-7765(02)00144-3 (2003).
Liu, J. H. et al. Role of flhDC in the expression of the nuclease gene nucA, cell division and flagellar synthesis in Serratia marcescens. J. Biomed. Sci. 7, 475–483, https://doi.org/10.1007/BF02253363 (2000).
Verstraeten, N. et al. Living on a surface: swarming and biofilm formation. Trends Microbiol. 16(496–506), 2008, https://doi.org/10.1016/j.tim.2008.07.004 (2008).
Aimin L, Tingteng L, Rong F (2007). Amidohydrolase superfamily. eLS. John Wiley & Sons, Hoboken, NJ.
Henderson, J. C. et al. The power of asymmetry: Architecture and assembly of the Gram-negative outer membrane lipid bilayer. Annu. Rev. Microbiol. 70, 255–78, https://doi.org/10.1146/annurev-micro-102215-095308 (2016).
Malinverni, J. C. & Silhavy, T. J. An ABC transport system that maintains lipid asymmetry in the Gram-negative outer membrane. Proc. Natl. Acad. Sci. USA 106, 8009–8014, https://doi.org/10.1073/pnas.0903229106. (2009).
Cabral, M. P. et al. Proteomic and functional analyses reveal a unique lifestyle for Acinetobacter baumannii biofilms and a key role for histidine metabolism. J. Proteome Res. 10, 3399–3417, https://doi.org/10.1021/pr101299j (2011).
Jang, I. A., Kim, J. & Park, W. Endogenous hydrogen peroxide increases biofilm formation by inducing exopolysaccharide production in Acinetobacter oleivorans DR1. Sci. Rep. 6, 21121, https://doi.org/10.1038/srep21121 (2016).
Srinivasan, R., Devi, K. R., Kannappan, A., Pandian, S. K. & Ravi, A. V. Piper betle and its bioactive metabolite phytol mitigates quorum sensing mediated virulence factors and biofilm of nosocomial pathogen Serratia marcescens in vitro. J. Ethnopharmacol. 193, 592–603, https://doi.org/10.1016/j.jep.2016.10.017 (2016).
King, J. D., Kocincova, D., Westman, E. L. & Lam, J. S. Review: lipopolysaccharide biosynthesis in Pseudomonas aeruginosa. Innate Immun. 15, 261–312, https://doi.org/10.1177/1753425909106436 (2009).
Beveridge, T. J. Structures of Gram-negative cell walls and their derived membrane vesicles. J. Bacteriol. 181, 4725–4733 (1999).
Zhang, X. S., García-Contreras, R. & Wood, T. K. YcfR (BhsA) influences Escherichia coli biofilm formation through stress response and surface hydrophobicity. J. Bacteriol. 189, 3051–3062, https://doi.org/10.1128/JB.01832-06, (2007).
Laport, M. S. & Lemos, J. A. Maria do Carmo, F. B., Burne, R. A. & Giambiagi-de Marval M. Transcriptional analysis of the groE and dnaK heat-shock operons of Enterococcus faecalis. Res. Microbiol. 155, 252–258, https://doi.org/10.1016/j.resmic.2004.02.002 (2004).
Khandekar, S. S., Daines, R. A. & Lonsdale, J. T. Bacterial β-ketoacyl-acyl carrier protein synthases as targets for antibacterial agents. Curr. Protein Pept. Sci. 4, 21–29, https://doi.org/10.2174/1389203033380377 (2003).
Taguchi et al. A homologue of the 3-oxoacyl-(acyl carrier protein) synthase III gene located in the glycosylation island of Pseudomonas syringae pv. tabaci regulates virulence factors via N-acyl homoserine lactone and fatty acid synthesis. J. Bacteriol. 188, 8376–8384, https://doi.org/10.1128/JB.00763-06 (2006).
Hoang, T. T., Sullivan, S. A., Cusick, J. K. & Schweizer, H. P. β-Ketoacyl acyl carrier protein reductase (FabG) activity of the fatty acid biosynthetic pathway is a determining factor of 3-oxo-homoserine lactone acyl chain lengths. Microbiology 148, 3849–3856, https://doi.org/10.1099/00221287-148-12-3849 (2002).
Vital-Lopez, F. G., Reifman, J. & Wallqvist, A. Biofilm formation mechanisms of Pseudomonas aeruginosa predicted via genome-scale kinetic models of bacterial metabolism. PLoS Comput Biol 11, e1004452, https://doi.org/10.1371/journal.pcbi.1004452 (2015).
Bernier, S. P., Letoffe, S., Delepierre, M. & Ghigo, J. M. Biogenic ammonia modifies antibiotic resistance at a distance in physically separated bacteria. Mol Microbiol 81, 705–716, https://doi.org/10.1111/j.1365-2958.2011.07724.x (2011).
Van Houdt, R., Moons, P., Buj, M. H. & Michiels, C. W. N-acyl-L-homoserine lactone quorum sensing controls butanediol fermentation in Serratia plymuthica RVH1 and Serratia marcescens MG1. J. Bacteriol. 188, 4570–4572, https://doi.org/10.1128/JB.00144-06 (2006).
Eitan, A. & Bibi, E. The core Escherichia coli signal recognition particle receptor contains only the N and G domains of FtsY. J. Bacteriol. 186, 2492–2494, https://doi.org/10.1128/JB.186.8.2492-2494.2004 (2004).
Jensen, L. J. et al. STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic acids research. 37(suppl_1), D412–D416, https://doi.org/10.1093/nar/gkn760 (2008).
Kamimura, N. et al. A bacterial aromatic aldehyde dehydrogenase critical for the efficient catabolism of syringaldehyde. Sci. Rep. 7, 44422, https://doi.org/10.1038/srep44422 (2017).
Anwar, N., Sem, X. H. & Rhen, M. Oxidoreductases that act as conditional virulence suppressors in Salmonella enterica serovar Typhimurium. PloS one 8, e64948, https://doi.org/10.1371/journal.pone.0064948 (2013).
Subramoni, S., Nguyen, D. T. & Sokol, P. A. Burkholderia cenocepacia ShvR-regulated genes that influence colony morphology, biofilm formation, and virulence. Infect. Immun. 79, 2984–2997, https://doi.org/10.1128/IAI.00170-11 (2011).
Palomino, M. M. et al. Influence of osmotic stress on the profile and gene expression of surface layer proteins in Lactobacillus acidophilus ATCC 4356. Appl. Microbiol. Biotechnol. 100, 8475–8484, https://doi.org/10.1007/s00253-016-7698-y (2016).
Bakkiyaraj, D., Sivasankar, C. & Pandian, S. K. Inhibition of quorum sensing regulated biofilm formation in Serratia marcescens causing nosocomial infections. Bioorg. Med Chem. Lett. 22, 3089–3094, https://doi.org/10.1016/j.bmcl.2012.03.063 (2012).
Annapoorani, A., Parameswari, R., Pandian, S. K. & Ravi., A. V. Methods to determine antipathogenic potential of phenolic and flavonoid compounds against urinary pathogen Serratia marcescens. J. Microbiol. Methods 91, 208–211, https://doi.org/10.1016/j.mimet.2012.06.007 (2012).
Nithya, C., Devi, M. G. & Pandian, S. K. A novel compound from the marine bacterium Bacillus pumilus S6-15 inhibits biofilm formation in Gram-positive and Gram-negative species. Biofouling 27, 519–528, https://doi.org/10.1080/08927014.2011.586127 (2011).
Yanhouy, L. et al. Antioxidant and glycation inhibitory activities of gold kiwifruit. Actinidia chinensis. J. Korean Soci. Appl. Biol. Chem. 54, 460–467, https://doi.org/10.3839/jksabc.2011.071 (2011).
Badireddy, A. R. et al. Spectroscopic characterization of extracellular polymeric substances from Escherichia coli and Serratia marcescens: suppression using sub-inhibitory concentrations of bismuth thiols. Biomacromolecules 9, 3079–3089, https://doi.org/10.1021/bm800600p (2008).
Durai, S., Vigneshwari, L. & Balamurugan, K. Caenorhabditis elegans‐based in vivo screening of bioactives from marine sponge‐associated bacteria against Vibrio alginolyticus. J. Appl. Microbiol. 115, 1329–422013, https://doi.org/10.1111/jam.12335 (2013).
Durai, S., Pandian, S. K. & Balamurugan, K. Establishment of a Caenorhabditis elegans infection model for Vibrio alginolyticus. J. Basic. Microbiol. 51, 243–252, https://doi.org/10.1002/jobm.201000303, (2011).
Sethupathy, S. et al. Proteomic analysis reveals modulation of iron homeostasis and oxidative stress response in Pseudomonas aeruginosa PAO1 by curcumin inhibiting quorum sensing regulated virulence factors and biofilm production. J. Proteomics 145, 112–126, https://doi.org/10.1016/j.jprot.2016.04.019, (2016).
Shevchenko, A., Tomas, H., Havli, J., Olsen, J. V. & Mann, M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856–2860, https://doi.org/10.1038/nprot.2006.468 (2006).
Ramos, S. et al. Effect of vancomycin on the proteome of the multiresistant Enterococcus faecium SU18strain. J. Proteomics. 113, 378–387, https://doi.org/10.1016/j.jprot.2014.10.012. (2015).
Acknowledgements
The authors are grateful for the computational facility provided by the Bioinformatics Infrastructure Facility funded by the Department of Biotechnology, Government of India [grant number BT/BI/25/015/2012(BIF)]. The Instrumentation Facility provided by Department of Science and Technology, Government of India through PURSE [Grant number SR/S9Z-3/2010/ 42(G)] and FIST [Grant number SR-FST/LSI-087/2008] and the University Grants Commission, New Delhi through SAP-DRS1 [Grant number F.3-28/2011 (SAP-II)] are gratefully acknowledged. The authors profusely thank Dr. Anton Hartmann, Professor, Research Unit Microbe-Plant Interactions, Helmholtz Zentrum München, Germany for providing the S. marcescens MG1.
Author information
Authors and Affiliations
Contributions
S.K.P., S.S., K.B. and S.M. conceived the experiments, S.S., S.A., A.S., B.S. and L.V. conducted the experiments, S.S., and S.K.P. wrote the main manuscript text and S.S. prepared the figures. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sethupathy, S., Ananthi, S., Selvaraj, A. et al. Vanillic acid from Actinidia deliciosa impedes virulence in Serratia marcescens by affecting S-layer, flagellin and fatty acid biosynthesis proteins. Sci Rep 7, 16328 (2017). https://doi.org/10.1038/s41598-017-16507-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-16507-x
This article is cited by
-
Sotolon is a natural virulence mitigating agent in Serratia marcescens
Archives of Microbiology (2021)
-
Global proteomic analysis deciphers the mechanism of action of plant derived oleic acid against Candida albicans virulence and biofilm formation
Scientific Reports (2020)
-
The upshot of Polyphenolic compounds on immunity amid COVID-19 pandemic and other emerging communicable diseases: An appraisal
Natural Products and Bioprospecting (2020)
-
Unraveling the anti-biofilm potential of green algal sulfated polysaccharides against Salmonella enterica and Vibrio harveyi
Applied Microbiology and Biotechnology (2020)
-
Quorum sensing inhibition and tobramycin acceleration in Chromobacterium violaceum by two natural cinnamic acid derivatives
Applied Microbiology and Biotechnology (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.