Abstract
The survival of wetland plant species largely relies on physiological adaptations essential for submergence and desiccation. Intertidal seaweeds, unlike terrestrial plants, have unique adaptations to submergence and can also sustain desiccation arising from tidal rhythms. This study determined the differential metabolic regulations in the inter-tidal seaweed species Ulva lactuca against the submergence and desiccation. During desiccation, the relative water content of the algal thalli declined with concomitant increase in reactive oxygen species (ROS) and lipid peroxidation. Nevertheless, the trends reversed during recovery on re-submergence and attained homeostasis. Metabolite profiling of U. lactuca revealed desiccation induced balance in energy reserve utilization by adjusting carbohydrate metabolism and switch over to ammonia metabolism. Upon re-submergence, thalli showed an increase in fermentative metabolites, pyruvate-alanine conversion, and the GABA shunt. Prolonged submergence induced substrate level phosphorylation mediated sugar biosynthesis while continuing the alternative carbon flux through fermentative metabolism, an increase in osmoprotectants glycine and betaine, sulfur bearing compounds cysteine and hypotaurine, and phenolic compound coniferaldehyde. The determined metabolic regulations in U. lactuca for submergence tolerance provide insights into potential evolutionarily conserved protective mechanisms across the green lineage and also highlights the possible role of sulfur oxoforms as strong free radical scavengers.
Similar content being viewed by others
Introduction
Marine macroalgae (seaweeds) represent the earliest diverging and evolutionary diverse aquatic organism occupying the basal position in the aquatic food web of intertidal and subtidal regions1. This group of aquatic plants have the unique physiology of being able to grow in fully submerged conditions representing the contrasting homeostasis regulations over land plants. Seaweeds, with their unique adaptability to submerged conditions make them a potential candidate to investigate the regulation of this trait. Seaweed species of the genus Ulva are most dominant in the upper intertidal zone where they regularly experience submergence and desiccation cycles with tides. The natural adaptation of Ulva species to submergence and desiccation therefore represents the existence of a unique homeostasis regulation. Furthermore, unlike terrestrial crops, Ulva has simple diastromatic thallus without any complex cellular level organization. This implies that the adaptive regulations are mostly cellular responses as opposed to complex anatomic modification in other plants to tolerate submergence. Earlier studies on understanding the homeostasis in Ulva were restricted to biochemical responses against the stress of irradiance2,3,4 salinity and temperature5 arising from desiccation in the intertidal region. The natural adaptability of this group of organisms to submergence is not been analysed.
In land plants, the prime downstream signalling regulator of submergence stress is ethylene biosynthesis6 which is presumed to be lost in marine plants such as seagrasses while their secondary adaptation to aquatic habitat7. This, therefore implies a unique metabolic regulations in marine plants or in seaweeds for maintaining their submerged physiology. The unavailability of extensive large-scale genomics data is the major bottleneck towards understanding the regulations in Ulva against acute stresses of intertidal zonation. Nevertheless, the differential expression of metabolites through untargeted metabolomics approach may provide insights into the possible underlying regulatory mechanisms.
The untargeted metabolomics may facilitate the functional annotations of a particular physiological state of an organism in the absence of whole genome information8. The metabolome analysis facilitates the determination of cell’s catalytic and regulatory processes. Further, metabolome represents the immediate biochemical consequences of genomic and transcriptomic activity and hence holds more biological relevance than other ‘-omes’9. In terrestrial life forms, metabolomics has made significant contributions towards investigating the responses to environmental stress, cues for biomarkers, chemotaxonomy, mutant differentiation and comparing growth stages, drug discovery, studying global effects of genetic manipulation, and natural product discovery10,11,12,13. Despite various advantages of metabolomics approach, studies are far behind in seaweeds compared to land plants. To date, no report describes the metabolic regulatory transitions happening during desiccation and submergence cycle in seaweeds. The present study is an attempt to understand the unique adaptation of Ulva lactuca tolerating periodic desiccation-submergence cycle using a holistic approach of untargeted metabolomics. Thus, a ‘learn from nature approach’ using a model organism naturally adapted for submergence and exposure, such as seaweed, can be supportive in understanding the submergence tolerance mechanisms either evolutionarily conserved or unique.
Results
Relative water content (RWC %)
Desiccation stress caused a significant decrease in the RWC. The RWC showed a marginal decline of 16% on desiccation exposure for 12 h which declined significantly with further increase in desiccation durations. The RWC declined to 28%, 33%, 38%, 42% and 58% in the thalli desiccated for 24, 36, 48, 72 and 96 h respectively (Fig. 1). On re-submergence after the desiccation time points of 12, 24, 36, 48 and 72 h, algae could able to recover and grow normally. This indicates that the RWC decline to less than 50% allow U. lactuca to recover on submergence. However, the thalli desiccated for 96 h lost more than 50% of RWC were not able to survive when rehydrated back in natural seawater.
Lipid peroxidation and ROS determination
The decrease in the RWC on desiccation was accompanied by an increase in lipid peroxidation and ROS generation in the thalli (Fig. 2A). The increase in lipid peroxidation was determined from an increased absorbance for malondialdehyde (MDA in nmol g−1 FW). The MDA accumulation showed a linear increase with the increase in desiccation duration. MDA level showed a slight increase of 2.21 ± 0.18 nmol g−1 FW after desiccation of 12 h. Prolonged desiccation for another 12 h showed two fold increase in lipid peroxidation (4.21 ± 0.21 nmol g−1 FW). Further extension in desiccation duration led to a significant increase in lipid peroxidation. The lipid peroxidation or MDA accumulation increased to 5.25 ± 0.24, 6.43 ± 0.24, 8.47 ± 0.13, 15.67 ± 0.48 nmol g−1 FW after desiccation of 36, 48, 72 and 96 h (Fig. 2A). Subsequent re-submergence reverted the lipid peroxidation levels to normal. The MDA accumulation on re-submergence was in the range of 2.08 ± 0.14 nmol g−1 FW to 3.41 ± 0.12 nmol g−1 FW for all the time points studied.
ROS generation was found to increase with an increase in desiccation duration (Fig. 2B). The ROS level (µmol g−1 FW) was found 0.05 ± 0.02 in control thalli while the ROS level showed ten-fold increase (0.15 ± 0.05 µmol g−1 FW) on desiccation for 12 h. The increase in desiccation duration to 24 and 36 h showed a steady increase in ROS levels to 0.20 ± 0.05, 0.28 ± 0.03 µmol g−1 FW. Further increase in desiccation period to 48, 72 and 96 h showed a linear-fold change in ROS generation with recorded values as 0.35 ± 0.05, 0.55 ± 0.04 and 0.68 ± 0.08 µmol g−1 FW respectively (Fig. 2B). On re-submergence for 2, 6 and 12 h after desiccation period of 12 h, the ROS level was estimated as 0.11 ± 0.05, 0.08 ± 0.04 and 0.07 ± 0.05 µmol g−1 FW respectively. The level of ROS remained in the range of 0.04 ± 0.02 to 0.08 ± 0.05 on continued submergence. The decline in ROS level from other desiccation time points were also in similar ranges as observed for desiccation of 12 h. This indicated the recovery potential of algal thalli on re-submergence after desiccation. However, algal thalli desiccated for 120 h on re-submergence did not show any change in ROS accumulation for 2 and 6 h and suddenly showed increase in ROS accumulation which led to decay in algal thalli. The histochemical staining employed to detect in situ accumulation of O2 •− and H2O2 radicals (two important representatives for ROS) using NBT and DAB respectively further confirmed the accumulation of ROS with the increase in desiccation followed by a decline in their accumulation on re-submergence. A blue formazone formed by reduction of NBT by O2 •− is clear evidence for the generation of superoxide radical (Fig. 2C). Similarly, the formation of H2O2 dependent brown precipitates was contingent with the exposure duration and accumulated maximum in the thalli exposed for longer duration.
From these analyses, samples desiccated for 24, 36 and 72 h and their subsequent submergence time points were selected for further analysis of metabolomics.
Metabolic profile
The untargeted metabolite profile for the seaweed species U. lactuca was generated using NMR spectroscopy. Metabolites identities were confirmed by both 1 H and 2D NMR approaches. The metabolite profile revealed the abundance of sugars, amino acids, organic acids, osmolytes, phenolic and sulfinic compounds (Table 1). The sugar metabolites identified were mainly the glucose and sucrose. The relative concentration of sucrose was highest among all the metabolites identified. Among amino acids, cysteine and glycine were high while other amino compounds such as choline, GABA, triethanolamine and glutamic acid were low. The fermentative pathway metabolites lactate, formic acid and acetate were also determined in this study. The compounds such as hypotaurine, ascorbate, allantoin, and coniferaldehyde were detected which have prominent anti-oxidative roles.
Metabolic variations during desiccation-submergence cycle
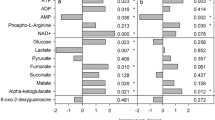
The metabolic variations were determined by comparing the normalized values of each metabolite detected at different treatment times. The samples of desiccation point of 24 h, 36 h and 72 h were compared with respect to always submerged control samples to determine the differential expression of metabolite during desiccation stress. All the metabolites identified showed significant correlation in their expression during different experimental conditions of desiccation and submergence (Fig. 3, Supplementary Figure 1). The results revealed a significant increase in all the metabolites except for sugars glucose, and sucrose after 24 h of desiccation. Further increase in desiccation to 36 h and 72 h showed contrasting behaviour with decline in the concentration of most of the identified metabolites except sugars glucose and sucrose (Fig. 4). The decline in the metabolite concentration was congruent to the increase in desiccation duration.
After determination of differential metabolite expression during desiccation, metabolite variations were determined in re-submerged samples (0.5 h, 1 h and 6 h) of each desiccation time point (Fig. 5). For this, the comparisons were initially made individually between the desiccation time point and its corresponding re-submergence time periods. The re-submergence of samples after a desiccation period of 24 h revealed a marked decline in the normalized values for all the metabolites detected at all the time points except for sucrose and allantoin (Fig. 5A). The algal thalli re-submerged after desiccation period of 36 h and 72 h showed contrasting metabolites expression compared to the same revealed after 24 h desiccation. The content of acetate, alanine, α-ketoglutaric acid, choline, GABA, glutamic acid, lactate and triethanolamine increased in both the formar cases while allantoin, and sucrose were decreased. Glucose and ascorbate showed contrasting behaviour in both the conditions with an increase on re-submergence after 36 h (Fig. 5B) of desiccation and decrease in the case of re-submergence after 72 h of desiccation (Fig. 5C). Other metabolites did not show significant differences. Further, a comparison of metabolite variation between each re-submergence time point was made. The results revealed increase in the normalized values for alanine, betaine, coniferaldehyde, and lactate, and decline in acetate, choline, GABA, glucose, glutamic acid, hypotaurine and sucrose, while no change in cysteine, glycine and triethanolamine at 0.5 h of re-submergence after desiccation period of 36 h and 72 h over the same after desiccation period of 24 h. Similar trends of metabolic variations were observed for 1 h and 6 h of re-submergence. However, small variations were observed on re-submergence for 6 h after desiccation period of 72 h whereas acetate and alanine declined while glucose, lactate and triethanolamine increased.
Discussion
The present study provides insights into the underlying metabolic regulations in intertidal seaweed species U. lactuca against the experienced desiccation and submergence. The cellular level damage by the excess production of ROS is the main source of injury. ROS generation is associated with most stresses of biotic and/or abiotic origin. Earlier studies on biochemical responses of desiccation stress tolerance in seaweeds revealed a marked increase in ROS generation14,15,16. The U. lactuca in this study could tolerate the RWC loss up to 50% with concomitant increase in ROS generation. The ROS generation and lipid peroxidation increased to the level that caused unrecoverable cellular damage when RWC loss exceeded above the 50%. The other intertidal seaweed species of genus Pyropia and/or Porphyra is reported to tolerate the water loss up to 90% despite overproduction of ROS and hypothesized to possess unique antioxidant machinery17. However, the non-tolerant seaweed species are reported to have major damage with slight misbalance in cellular water content18,19. Subsequent to desiccation, the re-submergence of algal thalli showed decline in ROS generation and lipid peroxidation confirming the recovery from the stress. Prolonged, submergence keep the ROS level stagnant which is in the limit of detoxification by normal antioxidative machinery.
Most of the compounds identified are the primary metabolites with known ubiquitous expression in land plants in different physiological conditions. Desiccation showed a significant decline in the concentrations of primary sugars glucose and sucrose reflecting the stress regulation mainly through increased carbohydrate metabolism18,19,20. The recorded decrease in primary sugars in the present study is associated with increase in α-ketoglutaric acid from TCA cycle, pyruvate mediated alanine biosynthesis and amine-group bearing compounds such as glycine, triethanolamine, and glutamic acid. Prolonged desiccation induced metabolic alterations to cease energy reserve by accumulation of sugars, TCA cycle compounds, and N metabolism while downregulating the energy expensive fermentative metabolic pathway compounds. The content of GABA also increased with increase in desiccation. Glycine, triethanolamine, and glutamic acid are regulators for glutathione biosynthesis and hence confirms their antioxidative role to scavenge the increased ROS with the increase in desiccation. Thus, the regulation against desiccation stress mainly includes the energy reserve depletion by increase in carbohydrate metabolism which in turn being switched over to ammonia metabolism through alanine, glutamine, and glutathione based regulatory pathways or photorespiration. Similar trends of metabolic variations were reported in E. siliculosus against the stress of hypo and hyper-salinity21. Accumulation of GABA is reported to be as a putative redox regulatory mechanism in various land plants like Lotus japonicus 5, Pisum sativum 22, Arabidopsis 23 and Glycine max 24.
The metabolic regulations for submergence tolerance showed important role of metabolites related to the pathways of glycolysis, fermentative and nitrogen metabolism. The increase in fermentative pathway metabolites i.e. acetate, lactate cause increased cellular acidification which is balanced by the increase in alanine, and activation of GABA shunt and glutamate-GABA cycle20. The presence of both acetate and lactate is indicative of the existence of PDH bypass which regenerates NAD+ and allows the tricarboxylic acid (TCA) cycle to continue25. Similar metabolic alterations have been reported from aquatic to land plants experiencing hypoxia on submergence22,23,24,25,26. On deprivation of carbon assimilation during submergence, the plants switch their metabolic activity to nitrogen assimilation27,28. This mechanism seems to be conserved and reported in unicellular primitive green alga Selenastrum minutum 29 to land crops such as Medicago 30 and rice31. The increase in glycine induce glutamate:glyoxylate aminotransferase activity was reported as regulators against hypoxia32,33,34. The same phenomenon is evidenced in the present study along with the activation of GABA shunt mediated through TCA cycle intermediate α-ketoglutaric acid. This supports an increases in ATP generation by proton gradient flux yielding energy under hypoxia. The results of the present study are in congruence to the reported hypoxia-induced metabolic regulations in seagrass species Zostera marina 26 and other plants32,33,34.
The metabolic alterations on submergence after 36 h and 72 h of desiccation were different from the same recorded for submergence after 24 h desiccation. The concentration of acetate, lactate, alanine, GABA and glutamic acid were higher in former two samples. Also, this increase in metabolites continues with the increase in time of submergence. The finding is in congruence to the metabolic regulations reported in higher plants that submergence increased metabolic flux towards fermentative and nitrogen assimilation pathways in response to applied environmental stresses. Interestingly, the rate of accumulation of primary metabolites of substrate level phoshorylation i.e. glucose along with osmoprotectants glycine and betaine were found to increase upon re-submergence after increase desiccation period. Our transcriptomics study revealed upregulation of S-adenosylmethionine synthetase (unpublished), a cofactor in the synthesis of compatible solutes such as glycine and betaine. This is also a major stress regulator from unicellular algae to complex land plant forms35,36. The increase in substrate level phosphorylation probably continues the ATP generation thus decrease the dependency over oxidative phosphorylation.
Interestingly, this study identified sulfur bearing metabolites cysteine and hypotaurine, phenolic compound coniferaldehyde with increased accumulation upon submergence. The former two are considered as non-proteinogenic cysteine-oxoforms having a promising role in free radicle detoxification37. The chemical structure of these metabolites make them labile for rapid transitions to different oxidative states eventually making them effective quenchers for free radicles8,9. Such metabolites along with cysteinsulfinic acid and isethionic acid were reported by Gupta et al.8,9 while performing untargeted metabolomics study in different seaweed species. However, their possible role was not determined. This study for the first time provide the evidence for their metabolic role in stress tolerance. A conceptualized model based on the findings from the present study is drawn to provide insights about the regulations seaweeds have to sustain and grow in submerge condition (Fig. 6).
Conclusion
This study for the first time revealed the differential metabolic regulations in intertidal seaweed species U. lactuca against desiccation-submergence cycle arising from periodic tidal rhythms. The desiccation stress is regulated by the balance in carbohydrate metabolism and switching to ammonia metabolism. The increase in glycolysis, fermentative and nitrogen metabolism is determined as major regulators to attain homeostasis upon submergence after desiccation. Also, the pyruvate-alanine cycle, and activation of GABA shunt increased with submergence. These regulations are conserved throughout the green lineage from unicellular to complex terrestrial plants. The prolonged submergence showed a distinct homeostatic response in U. lactuca with accumulation of more sugars along with osmoprotectants glycine and betaine. The fermentative metabolism continues indicating the NAD+ generation and the toxic effect of fermentative products is balanced by alternative carbon flux. Thus, seaweed have unique adaptation of energy generation during continuous submergence through substrate level phosphorylation, oxidative phosphorylation and controlled fermentative metabolism. The detailed transcriptome level study may further provide insights about the underlying regulations seaweed have for submergence tolerance unlike terrestrial plants.
Materials and Methods
Sample collection
Seaweed samples of species Ulva lactuca were collected from Veraval (N 20° 54.87′; E 70° 20.83′) along Gujarat coast, India. The collected thalli were maintained in unialgal culture in the laboratory in sterile enriched seawater media38 under white fluorescent lamps of irradiance intensity about 50 µmole photon m−2 s−1 with a 12: 12 h light:dark photoperiod. The algal thalli were made axenic following the method described by Reddy et al.39 and subjected to the stress of desiccation followed by submergence.
Desiccation and submergence treatment
For desiccation treatment, the thalli were exposed to air during light period while keeping their bottom surface in the wet sand (moisture content 10%). For this, the algal thalli were spread on a propylene plastic tray with autoclaved sea sand and incubated at 25 ± 2 °C with relative humidity 65 ± 2%. The desiccation treatment given was for different durations of 12, 24, 36, 48, 72, 96 h. Followed by each desiccation time, algal thalli were re-submerged in autoclaved seawater. After submergence, samples were collected at the periodic interval of 0.5, 1 and 6 h and analysed for their recovery by biochemical and metabolite characterizations.
Relative water content estimation
The relative water content (RWC) after the desiccation duration was considered as the measure to estimate the recovery of algal thalli on re-submergence. The RWC was calculated as follows: RWC = (LWt −DW)/(FW−DW) × 100, where LWt is the weight of the thalli subjected to the desiccation treatment for t hours, dry weight (DW) is the weight of the thalli oven dried for 48 h at 80 °C, and FW is the weight of the thalli before desiccation.
Determination of Reactive Oxygen Species (ROS)
ROS were determined according to the procedure described by Contreras et al.40. The thalli of U. lactuca (1 g FW) after different treatments of desiccation and submergence were incubated in 100 mL of 5 µM 2,4-dichlorofluoresceine diacetate (Calbiochem, San Diego, CA, USA) dissolved in filtered seawater for 1 h at 15 °C. After incubation, the thalli were rinsed in seawater, blotted dry, weighed, and frozen in liquid nitrogen. The tissues were then ground in liquid nitrogen, suspended in 5 mL of 40 mM Tris–HCl buffer, pH 7.0, and centrifuged at 8,000 × g for 25 min. Fluorescence was determined using LS-5 spectro-fluorometer (Perkin-Elmer, Norwalk, CT, USA) at an excitation wavelength of 488 nm and at an emission wavelength of 525 nm. Fluorescence values were obtained using a standard curve of 2,4-dichlorofluoresceine (DCF; Sigma, St. Louis, MO, USA).
Histochemical localization of O2 •− and H2O2
The production of O2 •− and H2O2 in response to desiccation and re-submergence was analyzed according to Castro-Mercado et al.41. For the detection of O2 •− radicals, small section of algal thalli (20 in number) each for control, desiccated and re-submerged algae were immersed in 5 mL detection solution containing 0.05% nitroblue tetrazolium (NBT) in 50 mM potassium phosphate buffer (pH 6.4) and 10 mM sodium azide (NaN3). The sections were infiltrated under vacuum for 3 min in the same solution and illuminated for 2 h until the appearance of dark spots, characteristic of blue formazan precipitates. Stained sections were cleared by boiling in acetic:glycerol:ethanol (1:1:3, v/v/v) solution before photographs were taken.
H2O2 production was visually detected by an endogenous peroxidase-dependent staining procedure using 3,3-diaminobenzidine (DAB). Algal thalli (20 in number each) were immersed in DAB solution 1 mg/mL (pH 5.0), vacuum-infiltrated for 5 min and then incubated at room temperature for 8 h in the presence of light till brown spots appeared. Sectioned were bleached by immersing in boiling ethanol to visualize the brown spots and photographs were taken.
Determination of lipid peroxidation
The level of lipid peroxidation in the thallus was determined as described by Heath and Packer42. The treated algal tissue (0.2 g) was extracted with 2 mL of 0.5% thiobarbituric acid (TBA) prepared in 20% trichloro acetic acid (TCA). The extract was heated at 95 °C for 30 min and then quickly cooled on ice. After centrifugation at 8,000 × g for 10 min, the absorbance of the supernatant was measured at 532 nm. Correction of non-specific turbidity was made by subtracting the absorbance value taken at 600 nm. The level of lipid peroxidation was expressed as nmol of malondialdehyde (MDA) formed using an extinction coefficient of 155 mM cm−1.
Metabolomics using NMR spectroscopy
The untargeted metabolite profiling was performed using NMR spectroscopy following the method developed by Gupta et al.9. The aqueous extract of algal thalli after treatments of desiccation and submergence were prepared. The algal thalli were blotted on paper towel to remove excess saline water and then thalli of 200 mg fresh wt. were macerated using mortar and pestle. While maceration about 200 µL of 50 mM phosphate buffer adjusted to pH 6.0 was added. The aqueous extracts were vortexed for one minute and then sonicated for 30 min with heating at 55 °C. The extracts were then centrifuged at 10,000 rpm for 2 min and clear supernatant was collected. An aliquot of this solution was transferred directly to 5 mm NMR tube to which a few drops of D2O containing a reference standard (TSP) was added.
NMR measurements and metabolite identification
NMR spectra were acquired on a Bruker Avance II 500 MHz spectrometer, equipped with a 5 mm BBI probe. Samples were spun at 20 Hz at temperature 20 °C. Each spectra was consisted of 80 scans of 2 s acquisition time with a spectral width of 7000 Hz. Spectra were Fourier transformed using an exponential line broadening value of 0.3 Hz. Spectra were then manually phased, baseline corrected and calibrated to the internal standard (TSP = 0.0 ppm). The solvent signal suppression was applied at 4.8 ppm during the recycle delay of 1 s with a low strength (bandwidth of 49 Hz) RF pulse. Ambiguities among the 1D pattern recognition were resolved by mapping the correlation among cross peaks of TOCSY (pre saturation version) and HSQC experiments. HSQC spectra were recorded by echo-antiecho method using gradients where 64 scans for 256 t1 points were acquired with recycle delay of 1.5 s. The total acquisition time was around 7 hrs. The broad residual water signal in HSQC spectra was removed by correcting the baseline with Gaussian function of width 0.3 ppm. TOCSY spectra were acquired with 24 scans for 128 t1 points. Metabolite identification was performed by comparing1H resonances and 2D correlation data against the reported literature and available online databases (http://prime.psc.riken.jp/, http://www.bmrb.wisc.edu/metabolomics/, http://www.hmdb.ca/), and Chenomx software (evaluation version).
Data processing
The spectral data hereby generated was examined for relative intensity variations among the identified metabolites by scaling the peak intensity. NMR peaks were bucketed in the range of 0.0–10.0 ppm excluding the water region from 4.5–5.0 ppm using MestReNova (Mestrelab Research, Spain). The spectral peaks were then sum normalized for scaling.
Statistical Analysis
All the experiments except NMR were performed with three biological replicates. The NMR experiments were carried out with two biological replicates. The data hereby presented is mean ± standard deviation (SD). The auto-scaled and log transformed metabolite profiles were analysed using the R and Bioconductor project43. Significantly present metabolites were selected using the significance level P < 0.05 as the cut-off after application of FDR correction for multiple testing. The heatmaps were plotted using pheatmap package of R.
References
Hurd, C. L., Harrison, P. J., Bischof, K. & Lobban, S. C. (eds.) Seaweed Ecology and Physiology, 2nd edn. Cambridge University Press, Cambridge, England (1994).
Dong, M., Zhang, X. & Zhuang, Z. et al. Characterization of the LhcSR gene under light and temperature stress in the green alga Ulva linza. Plant Mol. Biol. Rep. 30, 10–16 (2012).
Zhang, X., Cao, S. & Li, Y. et al. Expression of three putative early light-induced genes under different stress conditions in the green alga Ulva linza. Plant Mol. Biol. Rep. 30, 940–948 (2012).
Guan, Z. et al. Identification and expression analysis of four light harvesting-like (Lhc) genes associated with light and desiccation stress in Ulva linza. J. Exp. Mar. Biol. Ecol. 478, 10–15 (2016).
Kakinuma, M. et al. Physiological and biochemical responses to thermal and salinity stresses in a sterile mutant of Ulva pertusa (Ulvales, Chlorophyta). Mar Biol. 149, 97–106 (2006).
Voesenek, L. A. C. J. & Bailey-Serres, J. Flooding tolerance: O2 sensing and survival strategies. Curr. Opin. Plant Biol. 16, 647–653 (2013).
Golicz, A. A., Schliep, M. & Lee, H. T. et al. Genome-wide survey of the seagrass Zostera muelleri suggests modification of the ethylene signalling network. J. Exp. Bot. 66, 1489–1498 (2015).
Gupta, V., Thakur, R. S., Baghel, R. S., Reddy, C. R. K. & Jha, B. Seaweed metabolomics: a new facet of functional genomics. Adv. Bot. Res. 71, 31–52 (2014).
Gupta, V., Thakur, R. S., Reddy, C. R. K. & Jha, B. Central metabolic processes of marine macrophytic algae revealed from NMR based metabolome analysis. RSC Adv. 3, 7037–7047 (2013).
Higashi, Y. & Saito, K. Network analysis for gene discovery in plant-specialized metabolism. Plant Cell Environ. 36, 1597–1606 (2013).
Putri, S. P. et al. Current metabolomics: practical applications. J. Biosci. Bioeng. 115, 579–589 (2013).
Kim, H. K., Choi, Y. H. & Verpoorte, R. NMR-based plant metabolomics: where do we stand, where do we go? Trends Biotechnol. 29, 267–275 (2011).
Muranaka, T. & Saito, K. Phytochemical genomics on the way. Plant Cell Physiol. 54, 645–646 (2013).
Kumar, M. et al. Desiccation induced oxidative stress and its biochemical responses in intertidal red alga Gracilaria corticata (Gracilariales, Rhodophyta). Environ. Exp. Bot. 72, 194–201 (2011).
Flores–Molina, M. R. et al. Desiccation stress in intertidal seaweeds: effects on morphology, photosynthetic performance and antioxidant responses. Aquat. Bot. 113, 90–99 (2014).
Guajardo, E., Correa, J. A. & Contreras–Porcia, L. Role of abscisic acid (ABA) in activating antioxidant tolerance responses to desiccation stress in intertidal seaweed species. Planta 243, 1–15 (2016).
Contreras–Porcia, L., Thomas, D., Flores, V. & Correa, J. A. Tolerance to oxidative stress induced by desiccation in Porphyra columbina (Bangiales, Rhodophyta). J. Exp. Bot. 62, 1815–1829 (2011).
Contreras–Porcia, L., López–Cristoffanini, C., Lovazzano, C. & Flores–Molina, M. R. et al. Differential gene expression in Pyropia columbina (Bangiales, Rhodophyta) under natural hydration and desiccation conditions. Lat. Am. J. Aquat. Res. 41, 933–958 (2013).
López–Cristoffanini, C. et al. Identification of proteins involved in the tolerance responses to desiccation stress in the red seaweed Pyropia orbicularis (Rhodophyta. Bangiales). Proteomics 15, 3954–3968 (2015).
Bailey-Serres, J. et al. Making sense of low oxygen sensing. Trends in Plant Sci. 17, 129–138 (2012).
Dittami, S. M. et al. Integrative analysis of metabolite and transcript abundance during the shortterm response to saline and oxidative stress in the brown alga Ectocarpus siliculosus. Plant Cell Environ. 34, 629–642 (2011).
Zabalza, A., Orcaray, L. & Igal, M. et al. Unravelling the role of fermentation in the mode of action of acetolactate synthase inhibitors by metabolic profiling. J. Plant Physiol. 168, 1568–1575 (2011).
van Dongen, J. T. et al. Transcript and metabolite profiling of the adaptive response to mild decreases in oxygen concentration in the roots of Arabidopsis plants. Ann. Bot. 103, 269–80 (2009).
Allen, D. K. & Young, J. D. Carbon and nitrogen provisions alter the metabolic flux in developing soybean embryos. Plant Physiol. 161, 1458–1475 (2013).
Rocha, M. et al. Glycolysis and the tricarboxylic acid cycle are linked by alanine aminotransferase during hypoxia induced by waterlogging of Lotus japonicus. Plant Physiol. 152, 1501–1513 (2010).
Hasler-Sheetal, H., Fragner, L., Holmer, M. & Weckwerth, W. Diurnal effects of anoxia on the metabolome of the seagrass Zostera marina. Metabolom. 11, 1208–1218 (2015).
Stitt, M. et al. Steps towards an integrated view of nitrogen metabolism. J. Exp. Bot. 53, 959–970 (2002).
Barding, G. A., Fukao, T., Béni, S., Bailey-Serres, J. & Larive, C. K. Differential metabolic regulation governed by the rice SUB1A gene during submergence stress and identification of alanylglycine by 1H NMR spectroscopy. J. Prot. Res. 11, 320–330 (2012).
Vanlerberghe, G. C., Joy, K. W. & Turpin, D. H. Anaerobic metabolism in the N-limited green-alga Selenastrum minutum. Plant Physiol. 95, 655–658 (1991).
Ricoult, C., Cliquet, J. B. & Limami, A. M. Stimulation of alanine aminotransferase (AlaAT) gene expression and alanine accumulationin embryo axis of the model legume Medicago truncatula contribute to anoxia stress tolerance. Physiol. Plantarum 123, 30–39 (2005).
Miro, B. & Ismail, A. M. Tolerance of anaerobic conditions caused by flooding during germination and early growth in rice (Oryza sativa L.). Front Plant Sci. 4, 1–18 (2013).
Ricoult, C., Echeverria, L. O., Cliquet, J. B. & Limami, A. M. Characterization of alanine aminotransferase (AlaAT) multigene family and hypoxic response in young seedlings of the model legume Medicago truncatula. J. Exp. Bot. 57, 3079–3089 (2006).
Limami, A. M. Adaptations of nitrogen metabolism to oxygen deprivation in plants (eds. van Dongen, J. T. & Licausi, F.), Low-oxygen stress in plants. Plant Cell Mono. Vienna, Austria: Springer, 21, 209–221 (2014).
Limami, A. M., Glévarec, G., Ricoult, C. & Cliquet, J.-B. & Planchet, E. Concerted modulation of alanine and glutamate metabolism in young Medicago truncatula seedlings under hypoxic stress. J. Exp. Bot. 59, 2325–2335 (2008).
López–Cristoffanini, C., Tellier, F., Otaíza, R., Correa, J. A. & Contreras–Porcia, L. Tolerance to air exposure: a feature driving the latitudinal distribution of two sibling kelp species. Bot. Mar. 56, 431–440 (2013).
Hoekstra, F. A., Golovina, E. A. & Buitink, J. Mechanisms of plant desiccation tolerance. Trends Plant Sci. 6, 431–438 (2001).
Lynett, P. T., Butts, K., Vaidya, V., Garrett, G. E. & Pratt, D. A. The mechanism of radical-trapping antioxidant activity of plant-derived thiosulfinates. Org. Biomol. Chem. 9, 3320–3330 (2011).
Provasoli, L. Media and products for the cultivation of marine algae (eds. Watanabe, B. S. & Hattori, A.) Culture and collection of algae, 63–75 (The Japanese Soc. Plant Physiol. Tokyo, 1968).
Reddy., C. R. K. et al. An improved enzyme preparation for rapid mass production of protoplasts as seed stock for aquaculture of macrophytic marine green algae. Aquaculture 260, 290–297 (2006).
Contreras, L., Mella, D., Moenne, A. & Correa, J. A. Differential responses to copper induced oxidative stress in the marine macroalgae Lessonia nigrescens and Scytosiphon lomentaria (Phaeophyceae). Aquat. Toxicol. 94, 94–102 (2009).
Castro-Mercado, E., Martinez-Diaz, Y., Roman-Tehandon, N. & Garcia-Pineda, E. Biochemical analysis of reactive oxygen species production and antioxidative responses in unripe avocado (Persea americana Mill var Hass) fruits in response to wounding. Protoplasma 235, 67–76 (2009).
Heath, R. L. & Packer, L. Photoperoxidation in isolated chloroplasts. 1. Kinetics and stoichiometry of fatty acid peroxidation. Arch. Biochem. Biophys. 125, 189–198 (1968).
Gentleman, R. C. et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 5, R80 (2004).
Acknowledgements
VG and HRK like to acknowledge the financial support received from INSPIRE Faculty Award, Department of Science and Technology, New Delhi, India. The financial support from the project GAP 3022 and additional infrastructural support received from the project PSC0206 at CSIR-National Institute of Oceanography is acknowledged. The CSIR-National Institute of Oceanography contribution number for this manuscript is 6134. Authors acknowledge Prof. Juliet Coates, School of Biological Sciences, Birmingham University, UK for English and grammatical corrections.
Author information
Authors and Affiliations
Contributions
V.G. conceived the idea, executed the experiments. V.G. and H.R.K. analyzed the data. H.R.K. performed the statistical validation and draw correlation among samples and metabolites. V.G. and H.R.K. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gupta, V., Kushwaha, H.R. Metabolic regulatory oscillations in intertidal green seaweed Ulva lactuca against tidal cycles. Sci Rep 7, 16430 (2017). https://doi.org/10.1038/s41598-017-15994-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-15994-2
This article is cited by
-
Tissues and industrial co-products formed during alginate extraction from Laminaria hyperborea provide different metabolite profiles depending on harvest season
Journal of Applied Phycology (2023)
-
Benzopyrene induces oxidative stress and increases expression and activities of antioxidant enzymes, and CYP450 and GST metabolizing enzymes in Ulva lactuca (Chlorophyta)
Planta (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.