Abstract
Nuclear Pore Complex (NPC) is of paramount importance for cellular processes since it is the unique gateway for molecular exchange through the nucleus. Unraveling the modifications of the NPC structure in response to physiological cues, also called nuclear pore plasticity, is key to the understanding of the selectivity of this molecular machinery. As a step towards this goal, we use the optical super-resolution microscopy method called direct Stochastic Optical Reconstruction Microscopy (dSTORM), to analyze oocyte development impact on the internal structure and large-scale organization of the NPC. Staining of the FG-Nups proteins and the gp210 proteins allowed us to pinpoint a decrease of the global diameter by measuring the mean diameter of the central channel and the luminal ring of the NPC via autocorrelation image processing. Moreover, by using an angular and radial density function we show that development of the Xenopus laevis oocyte is correlated with a progressive decrease of the density of NPC and an ordering on a square lattice.
Similar content being viewed by others
Introduction
The unique gateway for bidirectional molecular exchange between the nucleus and the cytoplasm of eukaryotic cells is a complex molecular machinery spread across the nuclear envelope and called Nuclear Pore Complex (NPC)1,2,3,4. Whereas its composition and size may vary between different species, its general structure and function appear to be strikingly conserved5,6. The NPC is a large 60 to 125 MDa complex, composed of at least 30 different proteins, termed nucleoporins which are each present in multiple copies7,8. Recent studies have shown that half of these proteins are involved in the scaffolding of the NPC and arranged in an eight-fold symmetrical structure, such as the gp210 proteins, whereas the rest of the proteins, among which are the so-called FG-Nups, are considered to be unstructured and highly fluctuating9,10. Due to its size (>100 nm), its complexity and its internal dynamics, the study of NPC remains an experimental and theoretical challenge.
In the past years new approaches have been developed to achieve a better understanding of the NPC structure in response to external cues. Integrative approaches combining electron microscopy, proteomics and other biophysical tools have led to localize most of the constituents of the complex10,11,12. Cryo-electron microscopy in conjunction with subtomogram averaging have produced a 3D structure with a resolution of the NPC scaffold approaching 2 nm9. More recently, optical super-resolution microscopy has also been used successfully to study the structure of the NPC with a high molecular specificity of the labeling under physiological conditions13,14,15,16. In particular the ring shape eight fold symmetry of the pore has been resolved17 and the relative positions of the components of the Y complex, which is the essential building block of the NPC, have been measured18.
The modifications of the nuclear pore structure, also called nuclear plasticity, have been assessed using electron microscopy19,20,21,22,23,24,25. Cryo-electron microscopy studies have shown the existence of natural variants and modifications of its structure9,26,27, while atomic force microscopy has demonstrated a reversible mechanical gating of the nuclear pore28,29,30,31.
Using dSTORM super-resolution microscopy32,33, we showed that Xenopus Laevis oocyte development impacts on the structure and the large scale organization of the NPCs. By following some relevant parameters during developmental process, such as the internal and external nuclear pores diameter or their organization, we highlight structural modifications of the NPCs.
Results and Discussions
dSTORM imaging of the NPCs of Xenopus oocyte nuclear envelope
We have used dSTORM super-resolution imaging to gain insight for the first time into the organization and structure of the NPCs during oocytes development of X. Laevis. Xenopus nuclear envelopes were extracted and labelled at different stages of oocyte development using a protocol modified from earlier studies34,35 in order to preserve the integrity of the membrane and to ensure adhesion of the envelope to the glass coverslip (see Materials and Methods). Membranes were labeled using either a fluorescent wheat germ agglutinin (WGA-Alexa647 or WGA-Alexa488), which has a high affinity for N-acetylglucosamine modifications of the nucleoporins present in the nuclear pore central channel36,37, and/or a primary antibody (AB) against the gp210 protein coupled to a secondary mouse AB with a fluorescent marker (Alexa647). Ultimate structure of the NPCs was visualized using a dSTORM microscope (Zeiss Elyra P.S.1) (Fig. 1). At least 8 samples were imaged for each condition and more than 3 images were acquired for each sample. Each reconstructed image encompassed the recording of more than 30 000 frames, with at least 1 000 000 discrete localizations per sequence. Typically, around 30 000 NPCs were visible in each image. Thus, the analysis of more than 300 000 nuclear pores for each condition enabled us to uncover very subtle changes in the measured parameters. On the reconstructed image, the center of mass for each individual nuclear pore central channel was automatically detected. The diameters of the NPC central channel and the luminal ring (Fig. 2F,G) as well as the NPC density (Fig. 2D) were calculated and near neighbor angles were measured (Fig. 3D–F). The average image of the nuclear pore central channel was computed by combining individual reconstructed images of the pores (N = 270 000). In order to check whether the WGA labelling was not overestimating the measured densities of nuclear pores, the membrane was double stained with labeled WGA and anti-gp210 antibody (Fig. 1B). The NPCs density was similar in both cases, the percentage of pores labeled by WGA only being less than 6%.
dSTORM imaging of the nuclear envelopes from X. laevis oocytes. (A) dSTORM image of a spread nuclear envelope labelled with anti-gp210 primary antibody and Alexa647 secondary antibody. (B) dSTORM image of a spread nuclear envelope labelled with WGA-Alexa488. Scale bar 500 nm. (C) dSTORM image of a spread nuclear envelope labelled with WGA-Alexa488 (green) and gp210-Alexa647 (magenta). (D) Zoom on the image (A). (E) Zoom on the image (B).
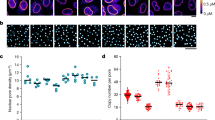
Effect of oocyte development on the density, the number and the diameter of the nuclear pore complexes. (A–C) dSTORM images of nuclear envelopes from oocytes respectively at stage II, IV and VI. The central channel is labelled with fluorescent WGA-Alexa647. Scale bar 5 µm. Insets: Stereomicroscope images of the oocyte respectively at stage II, IV and VI. Scale bar 50 µm. (D) Effect of oocyte development on nuclear pore complex density. (E) Effect of oocyte development on nuclear pore number per nucleus. (F) Effect of oocyte development on central channel diameter. (G) Effect of oocyte development on gp210 diameter. For each condition, the number of investigated NPC is superior to 300 000. Errors are experimental standard errors. The precision of the measurements was assessed by bootstrapping and by comparing different rounds of experiments.
Effect of oocytes development on the organization of the nuclear pore complexes. (A–C) 2D histogram of the probability P(d,α) to observe a NPC on a given envelope with two neighbors at a distance d and forming an angle α respectively for oocytes at stage II, IV and VI. For stage VI oocytes the most probable coordinates are (135 ± 5 nm, 90 ± 6°). The frequency event is normalized by the maximum for each histogram. (D–F) First neighbor angle distribution evolution for respectively stage II, stage IV and stage VI oocytes. The red dashed line at 60° corresponds to the minimal angle possible between 3 NPCs. (G) Experimental most probable angle α* between three nuclear pore complexes as a function of the distance d between the central pore and its two neighbors (dashed line) and theoretical angle α between three nuclear pore complexes as a function of the distance d between the central pore and its two neighbors in the packing model (solid line). In this model all the nuclear pore complexes are in contact with their first neighbors but without any large-scale order.
Effect of oocyte development on the density, number and diameter of the NPCs
Despite the fact that X. Laevis oocyte is one of the favorite model for the study of NPCs (see6 for a review), only two studies were carried out on early stage oocytes using electron microscopy38 or Atomic Force Microscopy (AFM)28. The other investigations involve late stage of oogenesis, i.e. stage VI39, because they are easier to handle due to their large size. As shown in Fig. 2D, the early stage II exhibit the highest density of NPCs with 53.8 ± 0.9 NPC.µm−2, while it was 46.2 ± 0.9 NPC.µm−2 at stage IV and 36.7 ± 0.8 NPC.µm−2 at stage VI (errors are experimental standard errors). The total number of NPCs per nucleus was computed from the diameter of the nuclei and nuclear pore density (Fig. 2E). Stage II oocytes had a lower number of NPCs (1.25 × 107) than stage IV oocytes (2.3 × 107) and stage VI oocytes (3.01 × 107). These results corroborate recent measurements carried out on native nuclear membranes from late stage oocytes by atomic force microscopy28 and super-resolution microscopy17 where the density of nuclear pores and the total number of NPCs per nucleus are in agreement with our observations. In contrast, these figures differ from the measurements obtained previously using negative staining methods and EM38. These discrepancies can be attributed to differences in sample preparation as discussed by Schlune et al.28 and come from the negative staining method which tends to select nuclear envelopes fragments with the highest pore frequency.
One of the major finding of our study is the observation of a decrease of the central channel and the scaffold diameter of the NPCs. Using optical super-resolution microscopy, which is compatible with specific labelling, we show a new structural plasticity of the NPCs at the level of the FG-Nups present in the central channel and at the level of the gp210 proteins involved in the scaffold of the NPCs. The NPCs of stage II oocytes had a central channel diameter of 40.8 ± 0.8 nm, at stage IV a diameter of 38.2 ± 0.8 nm (p(II-IV) < 0.01) and a diameter of 37.4 ± 0.8 nm at stage VI (p(II-VI) < 0.01). We can notice that the size of the central channel at stage VI is similar to that reported by optical super-resolution microscopy by Löschberger et al.17. In our case, the majority of the central channels of the NPCs is a compact structure without hole. This observation does not depend on the reconstruction parameters or the labelling protocol of the envelope (Figs S1 and S2). A similar compact structure has also been reported by STED super-resolution microscopy by Göttfert et al.14. For the luminal ring, we measure a diameter of 151.8 ± 1.8 nm for stage II oocytes, 148.6 ± 2.2 nm (p(II-IV) < 0.01) for stage IV and 144.3 ± 1.5 nm (p(II-VI) < 0.01) for stage VI (errors are experimental standard errors). Precision of all measurements was assessed by bootstrapping and by comparing different rounds of experiments40. Since previous studies show that transcriptional activity drops very significantly from a high level at early stage to an almost undetectable level at the late stage28,41, we argue that the dilatation may be linked to a direct mechanical effect such as the increase of the flow of matter like RNA circulating through the NPCs. Another explanation could be an allosteric switch in the structure of the pore as already observed for some of the FG-Nups present in the nuclear pore complex42 or other types of neuronal channels43,44,45. A separate study would be necessary to understand the physiological mechanism of this effect.
Effect of development stage on the lattice structure
The large-scale organization of the NPCs on the nuclear envelopes for stages II, IV and VI was determined by calculating the angle distribution between first neighbors for each NPCs and by using the angular and radial density function P(d,α). The latter describes the probability to observe on a given envelope a NPC with two neighbors at a distance d and forming an angle α. Qualitatively, we observed that the NPCs in earlier stages (II and IV) appeared to be less organized than in later stages (VI) as is it visible in Fig. 2A–C. Stage VI nuclear envelopes displayed small clusters of nuclear pores organized in square lattice with typical lateral size of 3 to 4 pores. In order to quantify this effect we measured the angle distribution function between the first two neighbors of a given NPC (Fig. 3D–F). For stage II and stage IV oocytes we can see a flat distribution which means that there are no preferential angles while at stage VI we have two distinct peaks at 90° and 180° (Fig. 3D–F). In order to investigate further this structure at all scales, different tools developed to analyze 2D and 3D crystal structure could be used such as higher order radial density function46 or bond-orientational order parameter47,48,49. We chose to use the angular and radial density function P(d,α) (Fig. 3A–C). This map can be seen as a measurement of the 2D order at all scales in the structure. A completely random structure will show a flat 2D map whereas a crystal-like structure will have a point like 2D map (Fig. S2). We observed two distinct behaviors for the early (II, IV) and late (VI) stages. In the case of stage II and stage IV we observed a map characteristic of a 2D dense amorphous structure. The maps could be divided into three regions: a low probability region corresponding to the close contact exclusion between discs of the same diameter, a high probability region corresponding to 2 neighbors in contact at a distance d and a flat region corresponding to non-overlapping NPC. In order to show that the high probability region corresponded to an amorphous organization of packed nuclear pores, we represented the most probable angle as a function of the distance between the NPC and its two neighbors. On the same axis we also represented the result of a simple packing model. In this model we assumed that the nuclear pores were closely packed. Then, the most probable angle α* formed by three NPC was determined by the contact at a distance d of the two other complexes which gives: sin α* = dNPC/d with dNPC the diameter of the complex. We observed a clear agreement between this model without any free parameter and the experimental data (Fig. 3G). This result showed that the structure at the early stages could be indeed described as a compact and amorphous structure of nuclear pore complexes without any particular order. In the case of later stage oocytes, we detected, in addition to the regions previously observed for early stages, a new localized high probability peak (d = 135 +/− 5 nm, α = 90 +/− 6°). This peak indicates the presence of neighbors positioned at the first neighbor distance and organized on a square lattice. This result showed quantitatively that the structure observed for late stage oocyte could be described as a mixture of amorphous and square lattice domains. These calculations demonstrate that the pores become organized and gradually form nano-domains with a square lattice structure on the nuclear envelope in the course of development. The first indication for a square lattice order in the NPCs organization in X. laevis oocyte has been provided by Unwin et al.38 for stage VI. Here we directly show that this order appears actually during oocyte development. Because it is already detectable at a low NPCs density, we propose that it is mediated by the gradual production of a structural constituent of the nuclear envelope. Lamins, which interact with the nuclear basket of the pore50,51,52,53,54, could be involved in this process55,56,57 and the molecular mechanism leading to this organization deserves further investigation.
Overall, we report for the first time that the oocyte development impacts on the nuclear organization and the structure of NPCs. Our studies could pave the way towards extensive works on how the structure of NPCs are linked with physiological activity and on the relation between the large scale organization of NPCs and the constituent of the nuclear envelope.
Materials and Methods
Living material
Xenopus laevis were purchased from “UMS 3387 - Centre de Ressources Biologiques Xénopes” and raised at 18 °C standard conditions in Xenoplus-amphibia housing system (Techniplast), illuminated with a photoperiod of 12 hours, in the animal housing facility at the Jacques Monod Institute (licence number B-75-13-17). Ovarian biopsies were performed on adult females anesthetized in 0.15% Tricaine methane sulfonate (MS222, Sigma Chemical, St. Louis, MO). Oocytes of different stages were sorted according to their distinct morphological features39 and incubated for 24 h at 18 °C in MBS Buffer (Modified Barth’s Solution). All animal experiments were performed in accordance with the approved protocols and guidelines at Paris Diderot University by the Animal Experimentation Ethical Committee Buffon (CEEA-40) supervised by the French ministry for education and research.
Nuclear envelope preparation and labelling
For dSTORM imaging of the nuclear envelopes of Xenopus oocytes at different stages, we used a protocol that is modified from Peters et al.35, Löschberger et al.17 and Penrad-Mobayed et al.34 in order to preserve the integrity of the membrane and to ensure the adhesion of the envelope with the glass coverslip even for early stages oocytes. Oocytes were transferred in 3:1 medium (75 mM KCl, 25 mM NaCl, Tris-HCl, pH 7.2). The nucleus is isolated by gentle pipetting after dissecting the oocyte using two pairs of forceps (Dumont N°5). The yolk granules still adhering to the nuclear envelope is removed by gentle backs and forth movement of the nucleus in the pipette. The nucleus is then transferred into a micro-chamber, filled with the same medium. The micro-chamber consists of a well with a circular hole (2.5 mm of diameter) in a glass slide, at the bottom of which a cover slide (Zeiss type 1.5, high precision, thickness 170 ± 5 µm) is sealed with cyanoacrylate glue. To open the small nucleus at early oocyte stages (approximately 250 μm of diameter for stage II), we used insect pins of 0.1 mm of diameter (Austerlitz stainless steel insect pins 0.1 mm) instead of the Dumont forceps, largely used for IV-VI oocyte stages. Furthermore, in order to make the nuclear envelope firmly attached to the cover slide before subsequent treatments, the nucleus is pressed to the bottom of the micro-chamber using blunt-end glass capillary. After dissection, the preparation is centrifuged at 300 g for 15 min, at 4 °C. After centrifugation, the samples were fixed for 20 min with 2% paraformaldehyde in phosphate-buffered saline (PBS), twice washed with PBS, saturated with 0.5% bovine serum albumin (BSA) for 10 min, labelled with fluorescent wheat germ agglutinin (WGA-Alex647, 1 µg.ml−1, Thermo Fisher Scientific) which is a lectin with a high affinity for N-acetylglucosamine modifications of the nucleoporins present in the nuclear pore central channel36,37. For the labelling of gp210, a mouse monclonal antibody against Xenopus gp210 kindly supplied by George Krohne (Wuerzburg University) have been used with the modified staining protocol of Löschberger et al.58, by adding a 5 min permeabilization step with 0.5% Triton-X100 between the fixation and the saturation steps. The preparations can be stored in PBS at 4 °C for 24–48 h before dSTORM imaging.
dSTORM imaging
All the dSTORM imaging was performed with a Zeiss Elyra P.S.1 microscope. In order to estimate the drift sample during imaging, we used either multicolor TetraSpeck beads (100 nm diameter, TetraSpeck; Life Technologies) as fiducial markers or either cross-correlation based algorithm from Zeiss-ZEN acquisition software. The photoswitching buffer was a commercial buffer from the Abbelight Company. The preparation was covered with a cover slide, sealed with a polymerized liquid (Twinsil Silicone, Rotec) and first observed under 10x air objective to look for the nuclear envelope. Then the region to be observed with dSTORM was selected with the 100x, NA 1.46, Zeiss plan-APO objective. With laser illumination at 642 nm and an emission filter matched to A647 emission spectrum (LP 655), we acquired 30000 raw images of blinking molecules, under total internal reflection fluorescence (TIRF) microscopy mode. The camera used was an EMCCD Andor iXon 897 (pixel size 16 µm, use of an Optovar lens magnification 1.6x, so the objective 100x used is yielding a final pixel size of 100 nm). Fluorophore positions were computed using the Zeiss-Zen super-localization software (ZEN). Briefly, the determination of the x-y coordinates of the fluorophores is achieved by approximation of a two-dimensional Gaussian function to the fluorescence emission pattern of individual spatially separated fluorophores in each frame. The localization precision of the x-y coordinates is calculated at the work of Mortensen et al.58. We then compensated the sample drift by tracking the immobile fluorescent beads or by using cross-correlation. Only molecules localized in the range of 10–30 nm were displayed, with a fluorescent signal of more than 500 detected photons per frame. The distribution of the typical relevant parameters such as the precision of localization or the full width at half maximum of the point spread function are displayed in Fig. S3. The color intensity is directly proportional to the localization density per pixel. The dSTORM images were reconstructed with a pixel size of 10 nm. The ZEN software outputs a text file containing a list of the x and y coordinates and the precision of all detected molecules in the time-series. We used this file to further visualize and treat the data with home-made Matlab program.
Image Analysis
The reconstructed super-resolution image is filtered using a hysteresis threshold. The center of mass for each individual nuclear pore central channels is detected from the regional maxima of the H-maxima transform. NPC density is computed from the number of centers of mass weighted by area occupied by the nuclear envelope on the image. The NPC central channel diameter is measured from the reconstructed image by interpolation of the full width at half maximum of the radial autocorrelation. The NPC luminal ring diameter is determined by interpolation of the third zero-value of the radial autocorrelation computed derivative. Angular and radial density function is computed from the local sampling of neighbor couples in a distance window [d − 35 nm, d + 35 nm] from the considered center of mass and then averaged on all the centers of mass.
Statistical test information
Alpha levels used in this work are 0.01. In other words we have considered the two tailed p-value tests significant for p < 0.01. The normality of our distributions has been tested by the evaluation of the first three moments of the distributions. We have used jack-knifing methods to assess that the standard error was a good estimate of the experimental error.
References
Pemberton, L., Blobel, G. & Rosenblum, J. Transport routes through the nuclear pore complex. Curr. Opin. Cell Biol. 10, 392–399 (1998).
Rout, M. P. & Blobel, G. Isolation of the yeast nuclear pore complex. J. Cell Biol. 123, 771–83 (1993).
Carmody, S. R. & Wente, S. R. mRNA nuclear export at a glance. J. Cell Sci. 122, 1933–7 (2009).
Tran, E. J. & Wente, S. R. Dynamic nuclear pore complexes: life on the edge. Cell 125, 1041–53 (2006).
Wente, S. R. & Rout, M. P. The Nuclear Pore Complex and Nuclear Transport. Cold Spring Harb. Perspect. Biol. https://doi.org/10.1101/cshperspect.a000562 (2010).
Floch, A. G., Palancade, B. & Doye, V. Fifty years of nuclear pores and nucleocytoplasmic transport studies: multiple tools revealing complex rules. Methods in cell biology 122 (2014).
Hoelz, A., Debler, E. W. & Blobel, G. The Structure of the Nuclear Pore Complex. in Annual Review of Biochemistry, Vol 80 (ed. Kornberg, R. D., Raetz, C. R. H., Rothman, J. E. & Thorner, J. W.) 80, 613–643 (Annual Reviews, 2011).
Grossman, E., Medalia, O. & Zwerger, M. Functional architecture of the nuclear pore complex. Annu. Rev. Biophys. 41, 557–84 (2012).
Eibauer, M. et al. Structure and gating of the nuclear pore complex. Nat. Commun. 6, 7532 (2015).
von Appen, A. et al. In situ structural analysis of the human nuclear pore complex. Nature 526, 140–143 (2015).
Alber, F. et al. Determining the architectures of macromolecular assemblies. Nature 450, 683–94 (2007).
Alber, F. et al. The molecular architecture of the nuclear pore complex. Nature 450, 695–701 (2007).
Schermelleh, L. et al. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science 320, 1332–6 (2008).
Göttfert, F. et al. Coaligned dual-channel STED nanoscopy and molecular diffusion analysis at 20 nm resolution. Biophys. J. 105, (2013).
Hüve, J., Wesselmann, R., Kahms, M. & Peters, R. 4Pi microscopy of the nuclear pore complex. Biophys. J. 95, 877–85 (2008).
Chatel, G., Desai, S. H., Mattheyses, A. L., Powers, M. A. & Fahrenkrog, B. Domain topology of nucleoporin Nup98 within the nuclear pore complex. J. Struct. Biol. 177, 81–89 (2012).
Löschberger, A. et al. Super-resolution imaging visualizes the eightfold symmetry of gp210 proteins around the nuclear pore complex and resolves the central channel with nanometer resolution. J. Cell Sci. 125, 570–5 (2012).
Szymborska, A. et al. Nuclear pore scaffold structure analyzed by super-resolution microscopy and particle averaging. Science 341, 655–8 (2013).
Goldberg, M. W., Wiese, C., Allen, T. D. & Wilson, K. L. Dimples, pores, star-rings, and thin rings on growing nuclear envelopes: evidence for structural intermediates in nuclear pore complex assembly. J. Cell Sci. 110(Pt 4), 409–20 (1997).
Kiseleva, E., Goldberg, M. W., Daneholt, B. & Allen, T. D. RNP export is mediated by structural reorganization of the nuclear pore basket. J. Mol. Biol. 260, 304–11 (1996).
Goldberg, M. W. & Allen, T. D. The nuclear pore complex and lamina: three-dimensional structures and interactions determined by field emission in-lens scanning electron microscopy. J. Mol. Biol. 257, 848–65 (1996).
Goldberg, M. W. & Allen, T. D. High resolution scanning electron microscopy of the nuclear envelope: Demonstration of a new, regular, fibrous lattice attached to the baskets of the nucleoplasmic face of the nuclear pores. J. Cell Biol. 119, 1429–1440 (1992).
Mehlin, H., Daneholt, B. & Skoglund, U. Translocation of a specific premessenger ribonucleoprotein particle through the nuclear pore studied with electron microscope tomography. Cell 69, 605–13 (1992).
Mehlin, H., Skoglund, U. & Daneholt, B. Transport of Balbiani ring granules through nuclear pores in Chironomus tentans. Exp Cell Res 193, 72–77 (1991).
Daneholt, B. A look at messenger RNP moving through the nuclear pore. Cell 88, 585–588 (1997).
Maimon, T., Elad, N., Dahan, I. & Medalia, O. The human nuclear pore complex as revealed by cryo-electron tomography. Struct. Lond. Engl. 1993(20), 998–1006 (2012).
Lim, R. Y. H., Aebi, U. & Fahrenkrog, B. Towards reconciling structure and function in the nuclear pore complex. Histochem. Cell Biol. 105–116 https://doi.org/10.1007/s00418-007-0371-x (2008).
Schlune, A., Shahin, V., Enss, K., Schillers, H. & Oberleithner, H. Plugs in nuclear pores: Transcripts in early oocyte development identified with nanotechniques. J. Cell. Biochem. 98, 567–576 (2006).
Shahin, V., Ludwig, Y. & Oberleithner, H. Investigation of nuclear envelope structure and passive permeability. Methods Mol. Biol. Clifton NJ 464, 161–80 (2009).
Liashkovich, I. et al. Clathrin inhibitor Pitstop-2 disrupts the nuclear pore complex permeability barrier. Sci. Rep. 5, 9994 (2015).
Liashkovich, I., Meyring, A., Oberleithner, H. & Shahin, V. Structural organization of the nuclear pore permeability barrier. J. Controlled Release 160, 601–608 (2012).
van de Linde, S. et al. Direct stochastic optical reconstruction microscopy with standard fluorescent probes. Nat. Protoc. 6, 991–1009 (2011).
Heilemann, M. et al. Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angew. Chem. Int. Ed Engl. 47, 6172–6 (2008).
Penrad-Mobayed, M., Kanhoush, R. & Perrin, C. Tips and tricks for preparing lampbrush chromosome spreads from Xenopus tropicalis oocytes. Methods San Diego Calif 51, 37–44 (2010).
Peters, R. Use of Xenopus laevis oocyte nuclei and nuclear envelopes in nucleocytoplasmic transport studies. Methods Mol. Biol. Clifton NJ 322, 259–72 (2006).
Davis, L. I. & Blobel, G. Nuclear pore complex contains a family of glycoproteins that includesp62: glycosylation through a previously unidentified cellular pathway. Proc. Natl. Acad. Sci. USA 84, 7552–6 (1987).
Finlay, D. R., Newmeyer, D. D., Price, T. M. & Forbes, D. J. Inhibition of invitro nuclear import by a lectin that binds to nuclear pores. J Cell Biol 104, 189–200 (1987).
Unwin, P. N. & Milligan, R. A. A large particle associated with the perimeter of the nuclear pore complex. J. Cell Biol. 93, 63–75 (1982).
Dumont, J. N. Oogenesis in Xenopus laevis (Daudin). I. Stages of oocyte development in laboratory maintained animals. J. Morphol. 136, 153–179 (1972).
Efron, B. Nonparametric estimates of standard error: The jackknife, the bootstrap and other methods. Biometrika 68, 589–599 (1981).
Sommerville, J. Using oocyte nuclei for studies on chromatin structure and gene expression. Methods San Diego Calif 51, 157–164 (2010).
Koh, J. & Blobel, G. Allosteric Regulation in Gating the Central Channel of the Nuclear Pore Complex. Cell 161, 1361–1373 (2016).
Corringer, P. J., Le Novere, N. & Changeux, J. P. Nicotinic receptors at the amino acid level. Annu. Rev. Pharmacol. Toxicol. 40, 431–458 (2000).
Paoletti, P. & Neyton, J. NMDA receptor subunits: function and pharmacology. Curr. Opin. Pharmacol. 7, 39–47 (2007).
Traynelis, S. F. et al. Glutamate Receptor Ion Channels: Structure, Regulation, and Function. Pharmacol. Rev. 62, 405–496 (2010).
Wochner, P. et al. X-ray cross correlation analysis uncovers hidden local symmetries in disordered matter. Proc. Natl. Acad. Sci. USA 106, 11511–4 (2009).
Jaster, A. The hexatic phase of the two-dimensional hard disk system. Phys. Lett. A 330, 120–125 (2004).
Marcus, A. H. & Rice, S. A. Observations of First-Order Liquid-to-Hexatic and Hexatic-to-Solid Phase Transitions in a Confined Colloid Suspension. Phys Rev Lett 77, 2577–2580 (1996).
Pindak, R., Moncton, D. E., Davey, S. C. & Goodby, J. W. X-Ray Observation of a Stacked Hexatic Liquid-Crystal B Phase. Phys Rev Lett 46, 1135–1138 (1981).
Smythe, C., Jenkins, H. E. & Hutchison, C. J. Incorporation of the nuclear pore basket protein nup153 into nuclear pore structures is dependent upon lamina assembly: evidence from cell-free extracts of Xenopus eggs. EMBO J. 19, 3918–3931 (2000).
Zhang, H., Saitoh, H. & Matunis, M. J. Enzymes of the SUMO modification pathway localize to filaments of the nuclear pore complex. Mol. Cell. Biol. 22, 6498–6508 (2002).
Liu, J. et al. Essential roles for Caenorhabditis elegans lamin gene in nuclear organization, cell cycle progression, and spatial organization of nuclear pore complexes. Mol. Biol. Cell 11, 3937–3947 (2000).
Lenz-Bohme, B. et al. Insertional mutation of the Drosophila nuclear lamin Dm0 gene results in defective nuclear envelopes, clustering of nuclear pore complexes, and accumulation of annulate lamellae. J. Cell Biol. 137, 1001–1016 (1997).
Mahamid, J. et al. Visualizing the molecular sociology at the HeLa cell nuclear periphery. Science 351, 969–972 (2016).
Maeshima, K. et al. Cell-cycle-dependent dynamics of nuclear pores: pore-free islands and lamins. J. Cell Sci. 119, 4442–4451 (2006).
Fiserova, J. & Goldberg, M. W. Relationships at the nuclear envelope: lamins and nuclear pore complexes in animals and plants. Biochem. Soc. Trans. 38, 829–831 (2010).
Aebi, U., Cohn, J., Buhle, L. & Gerace, L. The nuclear lamina is a meshwork of intermediate-type filaments. Nature 323, 560–564 (1986).
Mortensen, K. I., Stirling Churchman, L., Spudich, J. A. & Flyvbjerg, H. Optimized localization analysis for single-molecule tracking and super-resolution microscopy. Nat. Meth.
Acknowledgements
The authors want to thank Jean-Marc Di-Meglio, Guillaume Fabre, France Lam, Jean Antoine Lepesant, Valérie Doye, Catherine Dargemont, Caroline Perrin, Reiner Veitia for their help and discussions on this project, and Ludovic Maingault and Daniel Quintas for their care of the amphibians. The mouse monoclonal antibodies against Xenopus gp210 were kindly provided by Pr. Dr. George Krohne from Wuerzburg University. This project was supported by the Université Sorbonne Paris Cité (“NanoBioSTED” project grant), the Agence Nationale de la Recherche (ANR-14-CE08-0011-01, “Golden Gates” project grant) and the DIM Nano K (“RNA flow” project grant). This project was also supported by the French National Research Agency (ANR-10-INBS-04, FranceBioimaging, ≪ Investments of the future ≫).
Author information
Authors and Affiliations
Contributions
Conception and design of the experiments was done by O.F., M.P.-M., F.M. and J.S.L.A. was involved for the reagents supplying. The membrane preparation was carried out by J.S. and M.P.-M. J.S., C.G., A.F. and O.F. contributed to the dSTORM experiments. J.S. and F.M. performed the data analysis and developed the home-made MATLAB script. O.F., M.P.-M., F.M. and J.S. discussed the results, their presentation and wrote the manuscript. F.M. supervised the whole project.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sellés, J., Penrad-Mobayed, M., Guillaume, C. et al. Nuclear pore complex plasticity during developmental process as revealed by super-resolution microscopy. Sci Rep 7, 14732 (2017). https://doi.org/10.1038/s41598-017-15433-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-15433-2
This article is cited by
-
Percolation transition prescribes protein size-specific barrier to passive transport through the nuclear pore complex
Nature Communications (2022)
-
Nuclear Envelope and Nuclear Pore Complexes in Neurodegenerative Diseases—New Perspectives for Therapeutic Interventions
Molecular Neurobiology (2021)
-
Molecular architecture of the luminal ring of the Xenopus laevis nuclear pore complex
Cell Research (2020)
-
Deep learning massively accelerates super-resolution localization microscopy
Nature Biotechnology (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.