Abstract
The cornification of keratinocytes on the surface of skin and oral epithelia is associated with the degradation of nuclear DNA. The endonuclease DNase1L2 and the exonuclease Trex2 are expressed specifically in cornifying keratinocytes. Deletion of DNase1L2 causes retention of nuclear DNA in the tongue epithelium but not in the skin. Here we report that lack of Trex2 results in the accumulation of DNA fragments in the cytoplasm of cornifying lingual keratinocytes and co-deletion of DNase1L2 and Trex2 causes massive accumulation of DNA fragments throughout the cornified layers of the tongue epithelium. By contrast, cornification-associated DNA breakdown was not compromised in the epidermis. Aberrant retention of DNA in the tongue epithelium was associated neither with enhanced expression of DNA-driven response genes, such as Ifnb, Irf7 and Cxcl10, nor with inflammation. Of note, the expression of Tlr9, Aim2 and Tmem173, key DNA sensor genes, was markedly lower in keratinocytes and keratinocyte-built tissues than in macrophages and immune tissues, and DNA-driven response genes were not induced by introduction of DNA in keratinocytes. Altogether, our results indicate that DNase1L2 and Trex2 cooperate in the breakdown and degradation of DNA during cornification of lingual keratinocytes and aberrant DNA retention is tolerated in the oral epithelium.
Similar content being viewed by others
Introduction
Cornification is a mechanistically unique mode of programmed cell death that is restricted to keratinocytes undergoing terminal differentiation. Keratinocytes form multilayered squamous epithelia on the body surface and its invaginations such as the oral cavity. Within these epithelia, cells proliferate exclusively in the basal layer and undergo progressive differentiation in the suprabasal layers. Keratinocyte differentiation is associated with tightly regulated sequential changes in gene expression, modifications of proteins, and synthesis of particular lipids that build a physical barrier between the organism and the environment. Abnormalities in many cornification components result in enhanced transepidermal water loss, compromise the defence against infections, and underlie the development of several skin diseases. Distinct cornification programs give rise to the epidermis of the skin, oral and urogenital epithelia, and specialized skin appendages, such as hair, nails, and filiform papillae of the tongue, thereby conferring singular properties to each epithelium1,2,3.
Nuclear destruction is one of the most profound changes of the cell during cornification. Delay or absence of this breakdown process results in parakeratosis, which is found in a myriad of skin diseases, such as psoriasis and atopic dermatitis4, and also appears during wound healing5. While the appearance of parakeratosis in diverse skin disorders suggests that the degradation of the nucleus can be disturbed by multiple molecular triggers, the molecular regulation of cornification-associated breakdown of nuclear components and the etiological roles of these processes have remained poorly characterized. Phosphorylation of lamin A/C by Akt16, the nuclear transfer of profilaggrin fragments, and proteolytic activities of caspase-147,8,9 have been implicated in the control of keratinocyte enucleation.
The fragmentation and degradation of nuclear DNA are hallmarks of programmed cell death. Specific enzymes (DNases) degrade DNA in different physiological settings of cell death and, to a limited extent, during DNA repair10,11,12. DNases are also involved in the process of DNA degradation during cornification of keratinocytes. The keratinocyte-specific endonuclease DNase1L2 is required for nuclear DNA degradation in human keratinocytes differentiating in skin models in vitro 13. Knockout of DNase1L2 in the mouse revealed a crucial role of DNase1L2 in DNA degradation in terminally differentiated keratinocytes of hair, nails, tail scales, filiform papillae of the tongue, and in the esophagus14. Deletion of the lysosomal endonuclease DNase2 impaired DNA degradation during holocrine secretion of sebum but did not abrogate cornification-associated DNA breakdown in the interfollicular epidermis15,16. Only the deletion of both DNase1L2 and DNase2 blocked the removal of nuclear DNA during stratum corneum formation in the epidermis, suggesting that DNase1L2 and DNase2 cooperate in cornifying epidermal keratinocytes17. Trex2 is a 3′-exonuclease which is predominantly expressed in keratinocytes and contributes to the epidermal response to UVB-induced DNA damage18. Deficiency of Trex2 increased the susceptibility to skin carcinogenesis in mice18,19. Deletion of Trex2 also enhanced parakeratosis in the imiquimod-induced model of psoriasis in mice20. As the loss of Trex2 alone did not induce parakeratosis under tissue homeostasis but predisposed to parakeratosis under exposure to pro-inflammatory stress20, a cell-autonomous role of Trex2 in keratinocytes under non-inflammatory conditions remained elusive.
The blockade of DNA degradation by inactivation of the lysosomal endonuclease DNase211,21 or the loss of endonucleases responsible for extracellular DNA degradation, i.e. DNase122,23 and DNase1L324,25, results in the aberrant accumulation of self-DNA originating from dying cells, leading to over-activation of immune responses and subsequent onset of autoimmune diseases. Similarly, the loss of the exonuclease Trex1, which degrades DNA in the cytosol of many cell types, results in DNA accumulation leading to the induction of type I interferon (IFN) responses and autoimmunity26,27,28,29,30. The suppression of the nuclear DNA breakdown in cornifying keratinocytes does not induce skin inflammation14,16. Therefore, the lack of a tissue response to aberrantly retained DNA appears to be caused either by a suppressive mechanism that is associated with differentiated keratinocytes, such as Trex2-mediated degradation of DNA leaking into the cytosol, or by a putative lack of DNA-sensing and response factors in differentiated keratinocytes.
In the present study we investigated the effects of concomitant suppression of DNase1L2 and Trex2 on DNA degradation during cornification and on DNA-dependent inflammation. Taking into account that Trex2 processes DNA from 3′-OH ends31 that can be generated by DNase1-type nucleases, such as DNase1L213, we hypothesized that Trex2 and DNase1L2 might cooperate in DNA degradation. We demonstrate that there is an interplay between these two nucleases in lingual keratinocytes and accumulation of cytosolic DNA fragments in the absence of Trex2 and DNase1L2. Furthermore, our results show that homeostatic keratinocytes express only low levels of various DNA response genes and display tolerance to aberrantly retained endogenous DNA.
Results
The Trex2 exonuclease and the DNase1L2 endonuclease are expressed in differentiated keratinocytes
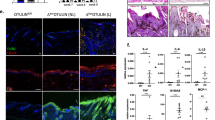
Among the multiple genes displaying DNA nuclease activity, the Trex2 exonuclease and the endonuclease DNase1L2 are specifically expressed in keratinocytes13,14,18,19. As shown in Fig. 1, immunofluorescence analysis revealed very similar patterns of expression of Trex2 and DNase1L2 proteins in differentiated keratinocytes of stratified squamous epithelia of both skin and tongue. The absence of staining in tissue samples from Trex2 and Dnase1l2 knockout mice confirmed the specificity of the immunolabelling reactions. Notably, the expression of both Trex2 and DNase1L2 proteins was increased in the upper keratinocyte layers, where endogenous DNA is degraded in the course of cornification. The distribution of DNase1L2 and Trex2 proteins in the epidermal layers correlated with the distribution of DNase1L2 and Trex2 mRNAs, that we obtained by analysis of the recently available transcriptomes of murine epidermal layers32. The mRNA levels of Trex2 as well as Dnase1l2 were highly upregulated specifically in the granular layer, whereas those of Trex1 and Dffb/CAD were downregulated, compared to their expression in the basal layer. Even though to less extend, the mRNA levels of nucleases Dnase1l3 and Dnase2a were also increased in the granular layer (Supplementary Figure S1). Consistently with previous studies33,34, epidermal differentiation was associated with increased expression of keratinocyte-specific Casp14 and decreased expression of the canonical apoptotic protease caspase-3 at the mRNA level (Supplementary Figure S1). Analysis of nuclease gene expression in available transcriptomes of wounded epithelia35 revealed that injury of the skin led to a decrease of the mRNA level of Dnase1l2 and to an increase of the expression levels of Trex2 whereas no changes were observed upon injury of the tongue (Supplementary Figure S2). These data suggest that the expression of DNase1L2 and Trex2 is regulated by different mechanisms in response to skin wounding but apparently in a similar manner in homeostatic epithelia of the skin and the tongue.
The nucleases Trex2 and DNase1L2 are mostly expressed in the keratinocyte upper layers of the skin and tongue epithelia. (a) Immunofluorescence images showing DNase1L2 (red) and DNA (blue) staining in the back skin and tongue sections from wild-type (wt) and Dnase1l2 knockout (Dnase1l2 −/−) mice. (b) Immunofluorescence images illustrating Trex2 (red) and DNA (blue) staining in back skin and tongue sections from wt and Trex2 knockout (Trex2 −/−) mice. Scale bars = 25 μm. Dashed lines, epidermal-dermal border; continuous lines, bottom of the stratum corneum; dotted lines, upper border of the stratum corneum.
Trex2 and DNase1l2 collaborate in the DNA degradation of lingual keratinocytes undergoing terminal differentiation
Fluorescent dye labelling of DNA (Fig. 1a and Fig. 2) and H&E staining (Fig. 2) showed that Dnase1l2 deficiency in mice led to retention of nuclei in the stratum corneum of the tongue (asterisks) but not of the skin, confirming our previous report14 that DNase1L2 was indispensable in the tongue epithelium whereas nuclear DNA degradation could proceed to completion even in the absence of DNase1L2 in cornifying epidermal keratinocytes. By contrast, DNA-specific fluorescent dye labelling showed that the loss of Trex2 did not result in aberrant DNA abundance in the stratum corneum of either skin18,19 or tongue under normal situations (Figs 1b and 2), suggesting that Trex2 action would be dispensable for most of nuclear DNA degradation. Nevertheless, this might partially be due to compensatory mechanisms.
Double Trex2 and Dnase1l2 deficiency does not trigger parakeratosis in the skin. H&E (lanes 1 and 2) staining and Hoechst DNA labelling (lanes 3 and 4) in back skin and tongue sections from wt, Trex2 −/−, Dnase1l2 −/− and Trex2 −/− Dnase1l2 −/− mice. Back skin samples were from 6 days-old mice which are consistently in the anagen phase of the hair cycle. Likewise, adult back skin was free from parakeratosis in all samples investigated. Tongue samples were from adult 7–9 weeks-old mice. The images are representative for at least five mice from each genotype. Scale bars = 25 μm. Dashed lines, epidermal-dermal border; continuous lines, bottom of the stratum corneum; dotted lines, upper border of the stratum corneum.
To determine whether the keratinocyte-specific Trex2 and DNase1L2 nuclease activities cooperate and/or overlap throughout the DNA degradation process leading to keratinocyte enucleation, we generated Trex2 Dnase1l2 (Trex2 −/− Dnase1l2 −/−) double knockout mice by crossing Trex2 (Trex2 −/−)18 and Dnase1l2 (Dnase1l2 −/−)14 knockout mice. The Trex2 −/− Dnase1l2 −/− mice were viable, born at the expected Mendelian frequency and indistinguishable by weight and growth from their double-heterozygous and single gene knockout littermates. These mice seemed healthy and no apparent skin abnormalities or an increase in skin diseases such as skin hyperpigmentation, alopecia, dryness and dermatitis, or spontaneous skin tumours were observed up to one year of age. Therefore, simultaneous loss of Trex2 and Dnase1l2 genes does not compromise survival into adulthood and does not generate an evident skin-related disease phenotype. Likewise the tongue was macroscopically inconspicuous in Trex2 −/− Dnase1l2 −/− mice. The numbers of living keratinocyte layers in the skin and lingual epithelia were not altered upon deletion of either Dnase1l2, Trex2 or both, suggesting that the inactivation of these DNases did not affect the cell turnover rates in epithelia.
To analyse the effects of the combined Trex2 and Dnase1l2 deficiencies on nuclear destruction during keratinocyte cornification, we first performed H&E staining and fluorescent labelling of DNA in skin and tongue samples from wild-type, Trex2 −/−, Dnase1l2 −/− and Trex2 −/− Dnase1l2 −/− mice. We did not detect abnormal retention of nuclei in the stratum corneum of interfollicular epidermis of back, ear and snout skin from Trex2 −/− Dnase1l2 −/−, Dnase1l2 −/− and Trex2 −/− mice by H&E staining and fluorescence labelling with a DNA-specific dye (Fig. 2, Supplemental Figure S3), indicating that neither single nor simultaneous loss of Trex2 and DNase1L2 prevented the removal of nuclei in cornifying epidermal keratinocytes. The lingual filiform papillae of Dnase1l2 −/− and Trex2 −/− Dnase1l2 −/− mice showed an aberrant accumulation of nuclear remnants in the cornified cell layers which was not observed in the corresponding samples of Trex2 −/− and wild-type mice (Fig. 2, asterisks), suggesting that loss of DNase1L214 but not loss of Trex2 leads to parakeratosis in lingual epithelia.
Next, we analysed the skin and tongue epithelia using the highly sensitive TUNEL assay for the presence of DNA fragments with free 3′-OH ends. In accordance with previous studies14,19,20, TUNEL-positive nuclei were observed sporadically in the epidermis of wild-type, Trex2 −/−, Dnase1l2 −/− and Trex2 −/− Dnase1l2 −/− mice. These nuclei were confined to epidermal keratinocytes in the transition stage from stratum granulosum to stratum corneum, indicating that DNA degradation during epidermal cornification involves TUNEL-positive intermediates both in the presence and absence of DNase1L2 and Trex2. In contrast to the uniform TUNEL labelling in the epidermis of all mouse strains investigated, there were profound differences in the TUNEL labelling in the cornified cell layers of the lingual stratum corneum of wild-type, Trex2 −/−, Dnase1l2 −/−, and Trex2 −/− Dnase1l2 −/− mice. TUNEL-positive nuclei were present in cornifying lingual keratinocytes of wild-type mice in a pattern similar to that observed in the epidermis (Fig. 3). Deletion of Trex2 resulted in the presence of TUNEL-positive DNA fragments not only in nucleus but also in the cytoplasm of lingual keratinocytes undergoing the transition from the living to cornified layer of the filiform papillae. In DNase1L2-deficient mice nuclear remnants within the parakeratotic filiform papillae were TUNEL-positive. The combined loss of Trex2 and Dnase1l2 was associated with the strong accumulation of fragmented DNA in the nucleus and in the cytosol of cornifying cells and in the entire stratum corneum (Fig. 3, panels in the third row). Lingual epithelium without filiform papillae displayed only sporadic TUNEL signals in wild-type, Trex2 −/−, and Dnase1l2 −/− mice but strong accumulation of TUNEL-positive DNA fragments, similar to that observed in filiform papillae, in Trex2 −/− Dnase1l2 −/− mice (Fig. 3, panels in the fourth row). These results indicated that (1) TUNEL-positive DNA fragments could be formed in a DNase1L2 and Trex2-independent manner during lingual keratinocyte cornification, and (2) Trex2 critically contributed to the degradation of nuclear DNA leaking into the cytosol of cornifying keratinocytes. Thus, cytosolic DNA fragments were confined to the transition cells of filiform papillae when only Trex2 was missing, suggesting that during normal cornification these fragments were reduced by Trex2 to levels below the detection limit of our assay. By contrast, when both Trex2 and DNase1L2 were absent, DNA fragments were either retained or further released from nuclear remnants when cornified cells moved towards the surface of the epithelium.
Double Trex2 and Dnase1l2 deficiency leads to an increase of fragmented DNA in the upper keratinocyte layers of the tongue, but not of the epidermis. TUNEL labelling of free 3′-OH DNA ends (red) and nuclei (blue) counterstained with Hoechst of back and ear skin, and the filiform papillae and back side tongue sections from wt, Trex2 −/−, Dnase1l2 −/− and Trex2 −/− Dnase1l2 −/− mice. Back and ear skin samples were from 6 days-old mice. Tongue samples were from 7–9 weeks-old mice. The images are representative for at least five mice from each genotype. Scale bars = 25 μm. Dashed lines, epidermal-dermal border; continuous lines, bottom of the stratum corneum; dotted lines, upper border of the stratum corneum.
Absence of immune response to aberrantly accumulated DNA in lingual differentiated keratinocytes
Intriguingly, in spite of the huge amount of cytosolic DNA aberrantly accumulated in the upper keratinocyte layers of the Trex2 −/− Dnase1l2 −/− tongue, no signs of inflammation and immune cell infiltration were histologically observed by H&E staining (Fig. 2), and no straight transcriptional signature of DNA-driven genes was found by RT-qPCR analysis (Fig. 4). In particular, although a significant, slight increase in the Mb21d1 (cGAS) mRNA expression levels was observed in Trex2 −/− Dnase1l2 −/− mice compared to wild-type mice, double deficiency of Trex2 and Dnase1l2 in the tongue did not induce major type I IFN and IFN-inducible genes, including Ifnβ, Irf7, Irf9, Mx1, Tlr9, and Aim2, or inflammatory genes, such as Tnf; mRNA levels of Cxcl10 were even lower in Trex2 −/− Dnase1l2 −/− mice than in wild-type mice. Furthermore, no significant changes in the mRNA levels of the keratinocyte differentiation genes Flg, Lor, Ivl and Krt10 were observed by single or double loss of the Trex2 and DNase1L2 nucleases (Supplementary Figure S4). Therefore, disrupted DNA degradation during lingual cornification does not result in the deregulation of the expression of key genes related to either DNA-driven responses or keratinocyte differentiation. Also, by indirect immunofluorescence analysis of antinuclear antibodies (ANAs), antibodies binding nuclear mitotic apparatus (NUMA) and nucleolus were detected in one out of five Trex2 −/− Dnase1l2 −/− mice and one out of five Dnase1l2 −/− mice, respectively (Supplementary Figure S5). Nevertheless, immuno-dot-blot analysis did not indicate increased serum levels of antibodies against chromatin antigens, such as nucleosomes and histones, in Dnase1l2 −/− or Trex2 −/− Dnasel12 −/− mice compared to Trex2 −/− and wild-type mice. Thus, single or double absence of keratinocyte-specific Trex2 and Dnase1l2 may trigger an immune response to nuclear-associated structures, but did not clearly result in DNA-driven immune response activation, even when defects in DNA degradation were evident.
DNA-driven gene expression signature is not induced in the tongue of the Trex2 −/− Dnase1l2 −/− mice. Expression of the indicated genes in the tongue from wt, Trex2 −/−, Dnase1l2 −/− and Trex2 −/− Dnase1l2 −/− mice, as determined by RT-qPCR. Each dot indicates a sample from an individual mouse, and horizontal lines represent the mean value. The results from one of two independent experiments with similar results are shown. Significant differences, by the Mann-Whitney test between genotypes: *P < 0.05; ***P < 0.001. AU, arbitrary units.
Because the activation of the type I IFN response relies primarily on nucleic acid recognition by innate immune sensors, we tested whether the unresponsiveness to DNA accumulation in lingual Trex2 −/− Dnase1l2 −/− keratinocytes could be associated with differences in the expression levels of DNA-driven sensing and signalling genes relative to cell types with robust DNA response. First, we investigated the AIM2 and cGAS-STING pathways, which are the major cytosolic DNA-sensing pathways known to be activated by endogenous cytosolic DNA36. As shown in Fig. 5, the mRNA levels of Aim2 and Tlr9, encoding key sensors for cytosolic and endosomal DNA respectively, and Tmem173 (STING), were more than 10-fold to 100-fold lower in tongue, skin and keratinocytes, as compared to immune tissues and cells, such as thymus, spleen and bone marrow-derived macrophages (BMDM). The mRNA expression levels of Mb21d1 (cGAS), a cytosolic DNA sensor upstream of STING, was hugely expressed in thymus compared to the rest of analysed samples revealing immune tissue-specific expression differences of key DNA signalling genes. STING was about 5-fold lower only in tongue but not in skin and keratinocytes, compared to spleen and BMDM. Notably, consistently with the lower inflammatory response observed in tongue compared to skin during wound healing35, the mRNA expression levels of these particular genes were lower in tongue than skin. In summary, at least Tlr9, Aim2 and STING, critical DNA sensing and signalling molecules, are expressed at relatively low levels in keratinocytes compared to BMDM, thereby likely intrinsically dampening DNA-driven responses in keratinocytes. Furthermore, consistent with the putatively reduced cell-intrinsic ability of keratinocytes to sense DNA and initiate a DNA-driven response, mouse keratinocytes were unresponsive to transfected DNA in comparison with BMDM (Fig. 6), similarly to what has been previously reported in human keratinocytes37. However, unlike human keratinocytes, TNFα stimulation did not render mouse keratinocyte responsive to cytosolic DNA (Fig. 6), thereby indicating another feature that distinguishes murine and human keratinocytes38. Taken together, our results suggest that mouse keratinocytes are intrinsically insensitive to cytosolic DNA, and this property may allow them to tolerate aberrant distribution of DNA in stratified epithelia such as that of the tongue.
DNA sensing genes are poorly expressed in keratinocytes compared to immune cells, and in skin compared to tongue. Expression of the indicated key DNA sensing genes in mouse thymus, spleen, tongue, skin, bone marrow-derived macrophages (BMDM) and keratinocytes, as determined by RT-qPCR. Graphs show mean and standard error of the mean (SEM) of at least three biological samples. AU, arbitrary units.
Intracellular DNA does not induce a transcriptional type I IFN response in mouse keratinocytes. Mouse bone marrow-derived macrophages (BMDM) and keratinocytes cells were untreated or treated with Lipofectamine® LTX (2 μl/ml), genomic DNA (2 μg/ml, premixed with Lipofectamine® LTX), LPS (10 ng/ml), TNFα (50 ng/ml) or a combination of these agents as indicated for 6 h prior to lysis. Total RNA was isolated, and mRNA levels of the indicated genes were quantified by RT-qPCR. Graphs show mean and SEM of at least three samples for each condition. The results from one of two independent experiments with similar results are shown. AU, arbitrary units.
Discussion
The present study reveals that two keratinocyte-specific nucleases, the exonuclease Trex2 and the endonuclease DNase1L2, cooperate in the DNA degradation process occurring during keratinocyte terminal differentiation in the tongue epithelium. As schematically depicted in Supplementary Figure S6, our data identifies Trex2 as the critical exonuclease that degrades DNA from the 3′-OH free ends that have been previously generated by DNase1L2. In vitro TREX2 activity studies have demonstrated that TREX2 acts specifically on 3′-OH DNA ends39,40. Furthermore, ectopic co-expression of TREX2 with rare-cleaving endonucleases has shown that TREX2 processing of 3′-ends alter non-homologous end joining double strand DNA repair pathways41,42,43. In the absence of DNase1L2, even though DNA is cleaved by so far undetermined nucleases, nuclear DNA remains present in corneocytes where it is mainly retained inside the nucleus. By contrast, loss of Trex2 results in the accumulation of fragmented DNA in the cytosol of cornifying keratinocytes. Taking into account that simultaneous loss of Trex2 and DNase1L2 leads to synergistic deposition of DNA fragments in the cytosol and the nuclei, rather than a simple sum of single DNase1L2 and Trex2 deficiency signatures, our results clearly point at these two nucleases as members of the same DNA degrading pathway in the tongue. Probably, DNA degradation by Trex2 exonuclease activity is partially overlapped or compensated by the DNA breakdown activity of DNase1L2 endonuclease. In fact, a slight increase in DNase1L2 expression, even though it did not reach statistical significance (P = 0.128), may occur in Trex2-deficient skin (Fig. 4) that could partially compensate Trex2 loss. Thus, only when the two gene activities are missing, the cooperative interplay among them becomes visible in the tongue under homeostatic conditions.
Of note, although Trex2 and DNase1L2 activities seem to be dispensable for homeostatic epidermal skin cornification, it cannot be ruled out that they contribute, individually or coordinately, to DNA removal during epidermal cornification under certain stress conditions and pathological situations such as cancer and inflammatory and autoimmune diseases. In this context, it has been shown that Trex2 itself plays a crucial role in DNA degradation during keratinocyte death either via apoptosis or cornification processes in the hyperplastic epidermis of both psoriatic and ultraviolet B-treated skin18,19,20. In fact, TREX2 seems to be the only DNase that is strongly upregulated in both human and mouse psoriasis forms20. DNase1L2 and DNase2 have been found to cooperate in the enucleation of skin corneocytes17, whereas DNase2 alone is critical for nuclear DNA degradation of sebocytes16. Altogether, these findings support tissue keratinocyte-specific relevance of particular DNases in the DNA degradation processes associated with distinct modes of keratinocyte terminal differentiation.
The differential tissue keratinocyte-type dependent actions of Trex2 and DNase1L2 may be relevant in DNA degradation processes within various contexts. The mRNA levels of other nucleases do not seem to be regulated upon injury and did not differ between tongue mucosa and skin. In addition, caspase-14, a differentiation-associated gene reportedly involved in keratinocyte nuclear destruction9,44,45 is only upregulated during wound healing in the skin but not in the tongue. Hence, the transcriptional responses of lingual and epidermal epithelia following injury are different. Oral and skin epithelia also display different susceptibilities to tumorigenesis46, reflecting relevant differences among tongue and skin epithelia and keratinocytes at either the levels of cell turnover, differentiation, morphology and/or metabolism. Future studies should determine whether Trex2 and DNase1L2 may play distinct roles in the tongue and skin keratinocytes under stress and pathological conditions, and their putative interplay with other nucleases and signalling pathways.
The distribution of Trex2 and DNase1L2 proteins, as determined by immunofluorescence, in epithelial layers parallel the spatial and temporal pattern of the loss of the nucleus in keratinocytes, supporting a crucial role of these nucleases in keratinocyte nuclear DNA degradation. At the mRNA level, the comparative analysis of basal, early spinous, late spinous and granular layer transcriptomes of murine epidermis32 uncovered that among known nucleases involved in intrinsic and extrinsic DNA degradation, Trex2 and Dnase1l2 were specifically and highly upregulated in the granular layer (Supplementary Figure S1), in agreement with their immunostaining patterns in skin epidermal layers (Fig. 1), while no major changes were observed in the mRNA abundance of Dffb/CAD, Dnase1, Dnase1l1, Dnase1l3, Dnase2a, and Trex1. Noteworthy, lingual cornification-associated DNA degradation can neither be compensated by ubiquitous Trex1, closely related to Trex231 and critically implicated in cytoplasmic DNA degradation47,48, nor Dffb/CAD, the main cell-autonomous DNase activated by caspase-3 that cleaves DNA in apoptotic cells49. Nevertheless, overlap and cooperation may take place, depending on tissue and pathways engaged to maintain homeostasis. For example, it has been proposed that DFFB/CAD is activated by caspase-14 and thus contributes to DNA degradation in differentiating human keratinocytes9. Caspase-14 is specifically upregulated (Supplementary Figure S1) and activated34,44,50,51 during keratinocyte differentiation and required for complete epidermis cornification, especially when its homeostasis is challenged7,45.
The absence of detectable DNA-driven immune responses against the huge accumulation of cytosolic self-DNA in the lingual epithelium of Trex2 −/− Dnase1l2 −/− mice, along with the mouse keratinocyte unresponsiveness to transfected DNA, as was also recently observed in human keratinocytes37, indicate that epithelia and keratinocytes, in particular, are highly tolerant to DNA. Accordingly, deletion of Trex1 in dendritic cells but not in keratinocytes leads to DNA-driven autoimmune phenotype in mice29. One explanation would be related to the level of expression of key DNA sensing and signalling genes. In line with this hypothesis, we showed that the mRNA expression amounts of Tmem173 (STING), which mediates type I IFN response to intracellular DNA52,53, were much lower in murine keratinocytes and keratinocyte-built epithelial tissues in tongue and skin, compared to macrophages and immune tissues. Consistently, Ifnβ and some key DNA-driven genes in the distinct keratinocyte layers were shown to be expressed at very low levels in mouse epidermal keratinocyte layers, and, with the notable exception of Irf7, were not transcriptionally induced (Aim2, Mb21d1, Mx1) or were downregulated (Cxcl10, Irf9, Tmem173) during keratinocyte differentiation in upper layers (Supplementary Figure S1). It has been suggested that low constitutive expression of type I IFN response genes in epithelial surfaces, which are naturally exposed to exogenous DNA, might be a strategy to avoid premature and excessive immune response54. Complementarily to the putatively low DNA-responsiveness of the keratinocytes, the lack of DNase1L2 and Trex2 may not only compromise degradation but also sensing of DNA in Trex2 −/− Dnase1l2 −/− tongue. In fact, Trex2 deficiency leads to attenuated skin inflammatory response to both imiquimod-induced psoriasis20 and ultraviolet-B radiation19. Furthermore, increasing evidence indicates supportive roles of distinct nucleases in nucleic acid immunity. The sensing of DNA by TLR9 requires digestion by DNase-II55, and short rather than large DNA fragments are efficient inflammasome activators56. Alternatively, immunostimulatory properties of DNA could be physically dampened as keratinocytes are committed to cornification. In particular, the formation of a cornified envelope may structurally prevent the release of DNA from terminally differentiated keratinocytes and, thus, its engulfment and sensing by dendritic cells and other immune cells that trigger potent immune responses to DNA. In contrast to keratinocytes, skin immune cells can be efficiently activated by endogenous nucleic acids, such as aberrant replicative DNA structures or damaged DNA, that may be released by highly proliferative or stressed keratinocytes and subsequently drive the onset and pathogenesis of skin inflammatory diseases, such as cutaneous lupus erythematosus57,58.
Prior to cornification-associated degradation of DNA, the organization of chromatin changes during terminal keratinocyte differentiation59,60. Thus it is conceivable that Trex2 and DNase1L2 mediate limited degradation of DNA during chromatin reorganization in differentiated keratinocytes when they are still alive. Potentially, such activities may influence gene expression during the final stages of keratinocyte differentiation. Interestingly, by means of mRNA sequencing, genes related to skin differentiation and chromatin biology, among others, are differentially expressed in Trex2 deficient imiquimod-treated skin as compared to treated wild-type skin20. Possible roles, besides DNA degradation in dying cells, of DNase1L2 and Trex2 deserve further studies.
In summary, we have shown that the Trex2 exonuclease and the DNase1L2 endonuclease, which are expressed in parallel to keratinocyte enucleation, cooperate and contribute to DNA degradation during keratinocyte terminal differentiation. Our demonstration of accumulating DNA fragments in the lingual epithelium of Trex2/DNase1L2-double deficient mice suggests not only a mechanism of DNA breakdown but, due to the absence of epithelial inflammation, also supports the concept of physiological tolerance of keratinocytes to intermediates of cornification-associated processing of endogenous DNA.
Methods
Mice
Trex2 −/− mice on C57BL/6 N background18 and Dnase1l2 −/− mice on 129SvEv14 have been described previously. The Trex2 −/− Dnase1l2 −/− mice were generated by crossing the Trex2 −/− and Dnase1l2 −/− mice. Dnase1l2 −/− mice and Trex2 −/− Dnase1l2 −/− mice were backcrossed eight times to C57BL/6 N background. The Trex2 −/−, Dnase1l2 −/−, Trex2 −/− Dnase1l2 −/− and wt C57BL/6 mice were housed and bred under specific pathogen-free environment in the animal facility of Parc Científic of Barcelona, University of Barcelona, according the institutional Ethics Committee on Animal Care. All animal experiments and methods were conducted under the authorizations of the Catalan Government (DAAM 7894 and DAAM 7895), and were performed in accordance with the relevant guidelines and regulations. Analyses were undertaken in 2 to 6-day old newborn mice, and 6- to 9-week old adult male and female mice, as indicated.
Histological and immunostaining analysis
Tissue samples from mice were fixed with 4% paraformaldehyde, embedded in paraffin and then cut in 5 μm sections. Sample processing for hematoxylin and eosin (H&E), staining and immunofluorescence assays was performed according to standard procedures33. For Trex2 immunofluorescence labelling, tissue sections were deparaffinised and rehydrated, and antigen retrieval was performed in Tris-EDTA buffer (pH 9.0). After blocking, sections were incubated with affinity-purified rabbit anti-mouse Trex2 (1/5018). Goat anti-rabbit IgG coupled to Alexa Fluor 555 (1/500; Life Technologies) was used as secondary antibody. Nuclei were counterstained with DAPI (Sigma), and sections were mounted in ProLong Gold Anti-Fade reagent (Life Technologies). For DNase1L2 immunofluorescence labelling, tissue sections were deparaffinised and rehydrated, and antigen retrieval was performed in citrate buffer pH6 (Dako). Sections were incubated with affinity-purified rabbit anti-DNase1L2 (1/10014). Goat anti-rabbit IgG coupled to Alexa Fluor 546 (1/500; Molecular Probes) was used as secondary antibody. Nuclei were counterstained with Hoechst 33258 (Sigma), and sections were mounted with Fluoprep (Biomerieux). Conventional images were captured using a Nikon Eclipse E-800 fluorescence microscope and an Olympus AX 70 microscope (Hamburg, Germany).
DNA labelling
The TUNEL (Terminal deoxynucleotidyl transferease dUTP nick end labeling) assay was performed to label 3′-OH ends of DNA in situ, according to the kit manufacturer’s instructions (In Situ Cell Death Detection Kit, TMR Red; Roche). Nuclear DNA was counterstained with Hoechst 33258 (Sigma). Slices were mounted onto glass slides using Fluoprep (bioMérieux, Marcy l′Etoile, France). Images were captured using an Olympus AX 70 microscope (Hamburg, Germany) equipped with a Spot RT3 slider camera (SPOT Imaging Solutions, Sterling Heights, MI).
Cells and treatments
Primary mouse keratinocyte cell cultures were prepared from 2 days-old newborn mouse skin and cultured in CNT-02 medium (CELLnTEC) for up to seven days, as previously described18, in either 24 or 12 wells plates. The medium was changed every second day. Cells were used for experiments at day 7 post plating. Bone marrow derived macrophages (BMDMs) from mice were cultured in DMEM supplemented with 20% fetal bovine serum and 30% supernatant of L929 cell conditioned media as a source of M-CSF, as previously described61. Macrophages were obtained as a homogeneous population of adherent cells after 7 days of culture. Then, cells were scrapped, counted and seeded for experiments in either 24 or 12-wells plates. Cells were left untreated, transfected with genomic DNA (2 μg/ml) using Lipofectamine® LTX (2 μl/ml; Invitrogen) or stimulated with 10 ng/ml of LPS (Sigma-Aldrich), 50 ng/ml TNFα (PreproTech Inc) or a combination of these agents, as indicated. Genomic DNA from mouse skin, treated with Proteinase K and RNase A, was purified using a silica-based column DNA isolation kit for tissues (Biotools B&M Labs), according to manufacturer’s indications, and eluted with water. For treatments, DNA was pre-incubated with Lipofectamine® LTX according to the manufacturer’s instructions. All cell treatments were left for 6 hours and then lysis was performed to extract total RNA.
RNA quantitative analysis
Total RNA from tissues and cells was isolated using the NucleoSpin RNA extraction kit (Macherey-Nagel), quantified and analysed using a NanoDrop ND-1000 Spectrophotomer, and cDNA synthesis was done using the GeneAmp RNA PCR kit (Applied Biosystems). Quantitative PCR (qPCR) amplification reactions were performed in the 7500 Fast Real-Time PCR System (Applied Biosystems). TaqMan Gene Expression Master Mix and predesigned probes (Applied Biosystems) were used to quantify mRNA expression of the following genes: Trex2 (Mm04210320_m1), Dnase1l2 (Mm00481868_g1), Ifnβ (Mm00439552_s1), Tnfα (Mm00443258_m1), Cxcl10 (Mm00445235_m1), Mx1 (Mm00487796_m1), Aim2 (Mm01295719_m1), IL6 (Mm00446190_m1), IL1β (Mm00434228_m1), and Sdha (Mm01352366_m1). Power SYBR Green PCR Master Mix (Applied Biosystems) and the following gene primers ere used to quantify mRNA expression of the following genes: Irf7 Forward (F) 5′GAGCAAGACCGTGTTTACG and Reverse (R) 5′CATGATGGTCACATCCAGG; Irf9 (F) 5′CAGTGTTCCTGGAGCATCAA and (R) 5′ACGCCTCTGTCAAGCTGATT; Tlr9 (F) 5′GAATCCTCCATCTCCCAACATG and (R) GAGGCTTCAGCTCACAGGG; Mb21d1 (F) 5′GTCGGAGTTCAAAGGTGTGGA and (R) 5′GACTCAGCGGATTTCCTCGTG; and Tmem173 (F) 5′CTACATTGGGTACTTGCGGTT and (R) 5′GCACCACTGAGCATGTTGTTATG. The results are presented relative to those for the housekeeping gene Sdha using the ΔCq method.
Supplementary Figures S1 and S2 show the processed transcriptomic data available from GSE23006 and GSE75931 respectively plotted in R.
Analysis of antinuclear antibodies
Human epithelial (HEp-2) cell substrates slides (INOVA Diagnostics) and goat anti-mouse IgG coupled to Alexa Fluor 488 (1/500; Life Technologies) was used to assess the presence of antinuclear antibodies (ANAs) in mouse sera, following the manufacturer’s instructions. Serum was obtained from blood that was extracted by maxillary punction from 9-weeks old mice, allowed to coagulate at room temperature, and then centrifuged. Supernatant was collected and stored at −80 °C for further ANA analysis. Immunofluorescence patterns were examined using a Nikon Eclipse E-800 fluorescence microscope at a magnification of 400X. An Immunodot Kit (BlueDot, D-tek) was used for the detection in mouse sera of IgG autoantibodies against nucleosomes and histones.
Statistical analysis
The Mann-Whitney test was applied for comparisons between genotypes. Test was two-tailed. The P values of ≤0.05 were considered to be statistically significant. Analysis was performed using GraphPad Prism software.
Data availability statement
The datasets generated and analysed during the current study are available from the corresponding authors on reasonable request.
References
Candi, E., Schmidt, R. & Melino, G. The cornified envelope: a model of cell death in the skin. Nat Rev Mol Cell Biol 6, 328–340, https://doi.org/10.1038/nrm1619 (2005).
Blanpain, C. & Fuchs, E. Epidermal homeostasis: a balancing act of stem cells in the skin. Nat Rev Mol Cell Biol 10, 207–217, https://doi.org/10.1038/nrm2636 (2009).
Eckhart, L., Lippens, S., Tschachler, E. & Declercq, W. Cell death by cornification. Biochim Biophys Acta 1833, 3471–3480, https://doi.org/10.1016/j.bbamcr.2013.06.010 (2013).
Cardoso, J. C. et al. ‘Hints’ in the horn: diagnostic clues in the stratum corneum. J Cutan Pathol 44, 256–278, https://doi.org/10.1111/cup.12839 (2017).
Usui, M. L., Underwood, R. A., Fleckman, P. & Olerud, J. E. Parakeratotic corneocytes play a unique role in human skin wound healing. J Invest Dermatol 133, 856–858, https://doi.org/10.1038/jid.2012.352 (2013).
Naeem, A. S., Zhu, Y., Di, W. L., Marmiroli, S. & O’Shaughnessy, R. F. AKT1-mediated Lamin A/C degradation is required for nuclear degradation and normal epidermal terminal differentiation. Cell Death Differ 22, 2123–2132, https://doi.org/10.1038/cdd.2015.62 (2015).
Hoste, E. et al. Caspase-14 is required for filaggrin degradation to natural moisturizing factors in the skin. J Invest Dermatol 131, 2233–2241, https://doi.org/10.1038/jid.2011.153 (2011).
Yamamoto-Tanaka, M. et al. Mesotrypsin and caspase-14 participate in prosaposin processing: potential relevance to epidermal permeability barrier formation. J Biol Chem 289, 20026–20038, https://doi.org/10.1074/jbc.M113.543421 (2014).
Yamamoto-Tanaka, M. et al. Multiple pathways are involved in DNA degradation during keratinocyte terminal differentiation. Cell Death Dis 5, e1181, https://doi.org/10.1038/cddis.2014.145 (2014).
Nagata, S. DNA degradation in development and programmed cell death. Annu Rev Immunol 23, 853–875, https://doi.org/10.1146/annurev.immunol.23.021704.115811 (2005).
Kawane, K., Motani, K. & Nagata, S. DNA degradation and its defects. Cold Spring Harb Perspect Biol 6, https://doi.org/10.1101/cshperspect.a016394 (2014).
Erdal, E., Haider, S., Rehwinkel, J., Harris, A. L. & McHugh, P. J. A prosurvival DNA damage-induced cytoplasmic interferon response is mediated by end resection factors and is limited by Trex1. Genes Dev 31, 353–369, https://doi.org/10.1101/gad.289769.116 (2017).
Fischer, H. et al. DNase1L2 degrades nuclear DNA during corneocyte formation. J Invest Dermatol 127, 24–30, https://doi.org/10.1038/sj.jid.5700503 (2007).
Fischer, H. et al. Essential role of the keratinocyte-specific endonuclease DNase1L2 in the removal of nuclear DNA from hair and nails. J Invest Dermatol 131, 1208–1215, https://doi.org/10.1038/jid.2011.13 (2011).
Fischer, H. et al. DNase 2 is the main DNA-degrading enzyme of the stratum corneum. PLoS One 6, e17581, https://doi.org/10.1371/journal.pone.0017581 (2011).
Fischer, H. et al. Holocrine Secretion of Sebum Is a Unique DNase2-Dependent Mode of Programmed Cell Death. J Invest Dermatol 137, 587–594, https://doi.org/10.1016/j.jid.2016.10.017 (2017).
Fischer, H., Buchberger, M., Napirei, M., Tschachler, E. & Eckhart, L. Inactivation of DNase1L2 and DNase2 in keratinocytes suppresses DNA degradation during epidermal cornification and results in constitutive parakeratosis. Sci Rep 7, 6433, https://doi.org/10.1038/s41598-017-06652-8 (2017).
Parra, D. et al. Increased Susceptibility to Skin Carcinogenesis in TREX2 Knockout Mice. Cancer Res 69, 6676–6684, https://doi.org/10.1158/0008-5472.CAN-09-1208 (2009).
Manils, J. et al. Multifaceted role of TREX2 in the skin defense against UV-induced skin carcinogenesis. Oncotarget 6, 22375–22396, https://doi.org/10.18632/oncotarget.4296 (2015).
Manils, J. et al. The Exonuclease Trex2 Shapes Psoriatic Phenotype. J Invest Dermatol 136, 2345–2355, https://doi.org/10.1016/j.jid.2016.05.122 (2016).
Kawane, K., Tanaka, H., Kitahara, Y., Shimaoka, S. & Nagata, S. Cytokine-dependent but acquired immunity-independent arthritis caused by DNA escaped from degradation. Proc Natl Acad Sci USA 107, 19432–19437, https://doi.org/10.1073/pnas.1010603107 (2010).
Napirei, M. et al. Features of systemic lupus erythematosus in Dnase1-deficient mice. Nat Genet 25, 177–181, https://doi.org/10.1038/76032 (2000).
Yasutomo, K. et al. Mutation of DNASE1 in people with systemic lupus erythematosus. Nat Genet 28, 313–314, https://doi.org/10.1038/91070 (2001).
Al-Mayouf, S. M. et al. Loss-of-function variant in DNASE1L3 causes a familial form of systemic lupus erythematosus. Nat Genet 43, 1186–1188, https://doi.org/10.1038/ng.975 (2011).
Sisirak, V. et al. Digestion of Chromatin in Apoptotic Cell Microparticles Prevents Autoimmunity. Cell 166, 88–101, https://doi.org/10.1016/j.cell.2016.05.034 (2016).
Morita, M. et al. Gene-targeted mice lacking the Trex1 (DNase III) 3′–>5′ DNA exonuclease develop inflammatory myocarditis. Mol Cell Biol 24, 6719–6727, https://doi.org/10.1128/MCB.24.15.6719-6727.2004 (2004).
Crow, Y. J. et al. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 cause Aicardi-Goutieres syndrome at the AGS1 locus. Nat Genet 38, 917–920, https://doi.org/10.1038/ng1845 (2006).
Lee-Kirsch, M. A. et al. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 are associated with systemic lupus erythematosus. Nat Genet 39, 1065–1067, https://doi.org/10.1038/ng2091 (2007).
Peschke, K. et al. Loss of Trex1 in Dendritic Cells Is Sufficient To Trigger Systemic Autoimmunity. J Immunol 197, 2157–2166, https://doi.org/10.4049/jimmunol.1600722 (2016).
Crow, Y. J. et al. Characterization of human disease phenotypes associated with mutations in TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, ADAR, and IFIH1. Am J Med Genet A 167A, 296–312, https://doi.org/10.1002/ajmg.a.36887 (2015).
Mazur, D. J. & Perrino, F. W. Identification and expression of the TREX1 and TREX2 cDNA sequences encoding mammalian 3′–>5′ exonucleases. J Biol Chem 274, 19655–19660 (1999).
Asare, A., Levorse, J. & Fuchs, E. Coupling organelle inheritance with mitosis to balance growth and differentiation. Science 355, doi:https://doi.org/10.1126/science.aah4701 (2017).
Fischer, H. et al. Caspase-14 but not caspase-3 is processed during the development of fetal mouse epidermis. Differentiation 73, 406–413, https://doi.org/10.1111/j.1432-0436.2005.00046.x (2005).
Lippens, S. et al. Epidermal differentiation does not involve the pro-apoptotic executioner caspases, but is associated with caspase-14 induction and processing. Cell Death Differ 7, 1218–1224, https://doi.org/10.1038/sj.cdd.4400785 (2000).
Chen, L. et al. Positional differences in the wound transcriptome of skin and oral mucosa. BMC Genomics 11, 471, https://doi.org/10.1186/1471-2164-11-471 (2010).
Roers, A., Hiller, B. & Hornung, V. Recognition of Endogenous Nucleic Acids by the Innate Immune System. Immunity 44, 739–754, https://doi.org/10.1016/j.immuni.2016.04.002 (2016).
Chiliveru, S. et al. Inflammatory cytokines break down intrinsic immunological tolerance of human primary keratinocytes to cytosolic DNA. J Immunol 192, 2395–2404, https://doi.org/10.4049/jimmunol.1302120 (2014).
Pasparakis, M., Haase, I. & Nestle, F. O. Mechanisms regulating skin immunity and inflammation. Nat Rev Immunol 14, 289–301, https://doi.org/10.1038/nri3646 (2014).
Mazur, D. J. & Perrino, F. W. Excision of 3′ termini by the Trex1 and TREX2 3′–>5′ exonucleases. Characterization of the recombinant proteins. J Biol Chem 276, 17022–17029 (2001).
Perrino, F. W., Harvey, S., McMillin, S. & Hollis, T. The human TREX2 3′ ->5′-exonuclease structure suggests a mechanism for efficient nonprocessive DNA catalysis. J Biol Chem 280, 15212–15218 (2005).
Bennardo, N., Gunn, A., Cheng, A., Hasty, P. & Stark, J. M. Limiting the persistence of a chromosome break diminishes its mutagenic potential. PLoS Genet 5, e1000683, https://doi.org/10.1371/journal.pgen.1000683 (2009).
Certo, M. T. et al. Coupling endonucleases with DNA end-processing enzymes to drive gene disruption. Nat Methods 9, 973–975, https://doi.org/10.1038/nmeth.2177 (2012).
Aubert, M. et al. In vitro Inactivation of Latent HSV by Targeted Mutagenesis Using an HSV-specific Homing Endonuclease. Mol Ther Nucleic Acids 3, e146, https://doi.org/10.1038/mtna.2013.75 (2014).
Denecker, G., Ovaere, P., Vandenabeele, P. & Declercq, W. Caspase-14 reveals its secrets. J Cell Biol 180, 451–458, https://doi.org/10.1083/jcb.200709098 (2008).
Hoste, E. et al. Caspase-14-deficient mice are more prone to the development of parakeratosis. J Invest Dermatol 133, 742–750, https://doi.org/10.1038/jid.2012.350 (2013).
Saiz-Ladera, C. et al. p21 suppresses inflammation and tumorigenesis on pRB-deficient stratified epithelia. Oncogene 33, 4599–4612, https://doi.org/10.1038/onc.2013.417 (2014).
Gehrke, N. et al. Oxidative damage of DNA confers resistance to cytosolic nuclease TREX1 degradation and potentiates STING-dependent immune sensing. Immunity 39, 482–495, https://doi.org/10.1016/j.immuni.2013.08.004 (2013).
Stetson, D. B., Ko, J. S., Heidmann, T. & Medzhitov, R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell 134, 587–598, https://doi.org/10.1016/j.cell.2008.06.032 (2008).
Samejima, K. & Earnshaw, W. C. Trashing the genome: the role of nucleases during apoptosis. Nat Rev Mol Cell Biol 6, 677–688, https://doi.org/10.1038/nrm1715 (2005).
Eckhart, L., Ban, J., Fischer, H. & Tschachler, E. Caspase-14: analysis of gene structure and mRNA expression during keratinocyte differentiation. Biochem Biophys Res Commun 277, 655–659, https://doi.org/10.1006/bbrc.2000.3698 (2000).
Fischer, H. et al. Stratum corneum-derived caspase-14 is catalytically active. FEBS Lett 577, 446–450, https://doi.org/10.1016/j.febslet.2004.10.046 (2004).
Ishikawa, H., Ma, Z. & Barber, G. N. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 461, 788–792, https://doi.org/10.1038/nature08476 (2009).
Paludan, S. R. Activation and regulation of DNA-driven immune responses. Microbiol Mol Biol Rev 79, 225–241, https://doi.org/10.1128/MMBR.00061-14 (2015).
Paludan, S. R. Innate Antiviral Defenses Independent of Inducible IFNalpha/beta Production. Trends Immunol 37, 588–596, https://doi.org/10.1016/j.it.2016.06.003 (2016).
Chan, M. P. et al. DNase II-dependent DNA digestion is required for DNA sensing by TLR9. Nat Commun 6, 5853, https://doi.org/10.1038/ncomms6853 (2015).
Dombrowski, Y. et al. Cytosolic DNA triggers inflammasome activation in keratinocytes in psoriatic lesions. Sci Transl Med 3, 82ra38, https://doi.org/10.1126/scitranslmed.3002001 (2011).
Peschke, K. et al. Deregulated type I IFN response in TREX1-associated familial chilblain lupus. J Invest Dermatol 134, 1456–1459, https://doi.org/10.1038/jid.2013.496 (2014).
Scholtissek, B. et al. Immunostimulatory Endogenous Nucleic Acids Drive the Lesional Inflammation in Cutaneous Lupus Erythematosus. J Invest Dermatol 137, 1484–1492, https://doi.org/10.1016/j.jid.2017.03.018 (2017).
Gdula, M. R. et al. Remodeling of three-dimensional organization of the nucleus during terminal keratinocyte differentiation in the epidermis. J Invest Dermatol 133, 2191–2201, https://doi.org/10.1038/jid.2013.66 (2013).
Botchkarev, V. A. The Molecular Revolution in Cutaneous Biology: Chromosomal Territories, Higher-Order Chromatin Remodeling, and the Control of Gene Expression in Keratinocytes. J Invest Dermatol 137, e93–e99, https://doi.org/10.1016/j.jid.2016.04.040 (2017).
Soler, C. et al. Macrophages require different nucleoside transport systems for proliferation and activation. FASEB J 15, 1979–1988, https://doi.org/10.1096/fj.01-0022com (2001).
Acknowledgements
We thank the Scientific and Technological Centers of the Universitat de Barcelona (Bellvitge) and Kinga Pajdzik and Sophie Bergmann (Medical University of Vienna) for their technical assistance. This study was supported by research grants from the Spanish Ministerio de Economía y Competitividad (BFU2010-15674 to CS, BFU2015-70581 to TV and SAF2014-55700-P to FC) and the Universitat de Barcelona (ACESB14 to CS). JM and EC were recipients of predoctoral fellowships from the Universitat de Barcelona and AGAUR, respectively.
Author information
Authors and Affiliations
Contributions
J.M., H.F., E.T., L.E. and C.S. designed the study; J.M., H.F., J.C., C.G., J.B., S.S., F.C., and J.M.d.A. performed research and analyzed data; E.C. and T.V. analyzed transcriptome data; C.S. and L.E. supervised the project, and J.M., H.F., E.T., L.E. and C.S. wrote the paper. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Manils, J., Fischer, H., Climent, J. et al. Double deficiency of Trex2 and DNase1L2 nucleases leads to accumulation of DNA in lingual cornifying keratinocytes without activating inflammatory responses. Sci Rep 7, 11902 (2017). https://doi.org/10.1038/s41598-017-12308-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-12308-4
This article is cited by
-
Multi-faceted attributes of salivary cell-free DNA as liquid biopsy biomarkers for gastric cancer detection
Biomarker Research (2023)
-
Single loss of a Trp53 allele triggers an increased oxidative, DNA damage and cytokine inflammatory responses through deregulation of IκBα expression
Cell Death & Disease (2021)
-
Cornification of nail keratinocytes requires autophagy for bulk degradation of intracellular proteins while sparing components of the cytoskeleton
Apoptosis (2019)
-
DNA methylation at an enhancer of the three prime repair exonuclease 2 gene (TREX2) is linked to gene expression and survival in laryngeal cancer
Clinical Epigenetics (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.