Abstract
Prenatal exposure to mercury, a known neurotoxic metal, is associated with lower cognitive performance during childhood. Disruption of fetal epigenetic programming could explain mercury’s neurodevelopmental effects. We screened for epigenome-wide methylation differences associated with maternal prenatal blood mercury levels in 321 cord blood DNA samples and examined the persistence of these alterations during early (n = 75; 2.9–4.9 years) and mid-childhood (n = 291; 6.7–10.5 years). Among males, prenatal mercury levels were associated with lower regional cord blood DNA methylation at the Paraoxonase 1 gene (PON1) that persisted in early childhood and was attenuated in mid-childhood blood. Cord blood methylation at the PON1 locus predicted lower cognitive test scores measured during early childhood. Methylation at the PON1 locus was associated with PON1 expression in an independent set of cord blood samples. The observed persistent epigenetic disruption of the PON1 gene may modulate mercury toxicity in humans and might serve as a biomarker of exposure and disease susceptibility.
Similar content being viewed by others
Introduction
Mercury (Hg) is a ubiquitous worldwide environmental contaminant that can persist in the environment and bioaccumulates as methylmercury (MeHg) in the food chain. Elemental mercury is a natural metal that is primarily introduced in the air, water and food from coal burning power plants, artisanal mining activities and industrial applications1. Since the industrial revolution, mercury levels in surface water have tripled2. For the general public, the consumption of fish or seafood containing MeHg is the major source of exposure3.
MeHg crosses the placenta and the blood-brain barrier and accumulates in fetal tissues resulting in fetal blood concentrations that typically exceed maternal levels4. Prenatal effects of acute MeHg exposure were first documented in 1956, when industrial activity heavily polluted Japan’s Minamata bay causing coastal residents to consume highly contaminated seafood. Some infants born during this period suffered from severe neurological disabilities and fetal abnormalities5. Recent prospective epidemiological studies have shown that even moderate or low-level prenatal exposure to mercury in utero typical of regular fish consumption may be associated with lower cognitive test scores in children6,7,8,9,10. However, the dose-response relationship, mechanism of action and potential effect modifiers such as gender and timing of exposure have not been fully evaluated11.
The specific mechanisms of neurotoxicity and cognitive disruption associated with prenatal mercury exposure remain poorly characterized in humans. Nonetheless, the period of fetal development has been shown to be sensitive to prenatal exposure, perhaps in part because of the dramatic DNA methylation changes and epigenomic remodeling that takes place early during embryogenesis, giving rise to cells and tissues with specific DNA methylation patterns12. The ability of mercury to cross the placenta and blood-brain barrier during development makes it a candidate toxicant for the disruption of fetal programming events that could propagate through different germ layers during embryogenesis.
Although several studies have characterized DNA methylation changes in cord blood relative to prenatal environmental exposures, very few studies have evaluated the persistence of these changes into childhood13. Evaluating the persistence of epigenetic modifications is critical as DNA methylation is a dynamic process that can drift with age and other stochastic processes14. To date, three epigenome-wide association studies have been conducted identifying unique genomic regions as well as individual CpG methylation disruption in cord blood15, 16 and placenta17 of newborns prenatally exposed to mercury. However, these studies were limited in sample size (ranging from 41 to 141 samples) and did not evaluate whether the observed epigenetic alterations were persistent, sex-specific or related to cognitive performance in childhood. Epigenetic responses to environmental conditions can be sex specific in terms of perception of exposure and subsequent response, particularly for neurodevelopmental disorders18, 19. Furthermore, gender differences in MeHg neurotoxicity have been documented in epidemiological and animal models20, 21. Therefore, it is crucial to evaluate sex-specific epigenetic alterations in response to environmental conditions.
In the present study, we examined associations of prenatal mercury exposure with epigenome wide DNA methylation in cord blood, and evaluated if these epigenetic modifications persist in blood through early and mid-childhood. We also evaluated sex-specific differences in response to prenatal mercury exposure. We hypothesized that persistent DNA methylation modifications in cord blood related to prenatal mercury exposure would also be associated with cognitive test scores measured in early childhood.
Results
A total of 321 mother-child pairs had available data on both maternal prenatal mercury exposure and cord blood DNA methylation. During early childhood (range: 2.9 to 4.9 years), 75 participants had whole blood samples available for analysis and 291 participants had samples available for mid-childhood analyses (range: 6.7 to 10.5 years), Table 1. Mean maternal red blood cell mercury (RBC-Hg) concentration measured during the second trimester of pregnancy was 3.8 ng/g (SD = 3.1) and mean maternal fish intake during pregnancy was 1.5 servings per week (SD = 1.2). Prenatal mercury concentration in maternal erythrocytes was right skewed and so we log2-transformed it for analyses to approximate a log-normal distribution, Supplementary Fig. S1. Univariate associations among the main 30 principal components of DNA methylation data, explaining 51.4% of the variance, showed strong associations with estimated cell type composition and moderate associations with phenotype characteristics after the effective removal of technical batch effects, Fig. 1.
Regional DNA Methylation Analysis
To identify Differentially Methylated Regions (DMRs) in the genome relative to prenatal mercury exposure, we implemented a regional based approach that is agnostic to both genomic annotation and direction of individual CpG associations. After stratifying on sex, a DMR covering 9 CpGs of chromosome 7 in the Paraoxonase 1 gene (PON1) was hypomethylated by prenatal mercury exposure among boys (genomic coordinates: chr7:94,953,653–94,954,202; FDR < 0.05), Fig. 2. We did not identify any DMRs in girls, or in the overall population. Among males, the multivariate adjusted magnitude of association for each individual CpG site in the PON1 DMR ranged from a 1% to a 3.8% decrease in individual cord blood CpG methylation per doubling in maternal RBC-Hg concentrations, Table 2. Overall, mean cord methylation levels of all nine CpGs located in the PON1 locus were inversely associated with maternal RBC-Hg concentrations (β = −2.4%, 95% CI: −3.8, −1.0; P = 7.5 × 10−4), Table 2. Scatter plots with locally weighted smoothing lines by median prenatal mercury concentrations levels of the PON1 DMR are shown in Fig. 3. Individual cord blood methylation levels for the 9 CpGs in this DMR ranged from 25.2% to 98.9% (Fig. 4A) and they were strongly positively correlated with each other (ρ-range: 0.72 to 0.96), Fig. 4B.
Cord blood CpG methylation distribution and fitted locally weighted scatterplot smoothing (LOESS) lines in the DMR found in PON1 (9 CpGs) with low (blue) and high (red) prenatal mercury exposure among males. Rug plot represents the estimated CpG density in the region while the green area denotes a CpG Island.
To evaluate the persistence of association between prenatal mercury exposure and DNA methylation at the PON1 locus found in cord blood, we tested associations between prenatal mercury exposure with DNA methylation at the PON1 locus in blood collected in males at early and mid-childhood. In early childhood (n = 37), a doubling in prenatal mercury concentration was associated with a 4.6% decrease in mean methylation levels of the DMR in PON1, measured across the nine CpGs (β = −4.6%, 95% CI: −9.0, −0.1; P = 0.044). Investigating all 9 CpGs individually, the inverse adjusted association of prenatal mercury exposure reached statistical significance in early childhood among five CpGs: cg07404485 (β = −4.2%, 95% CI: −7.8, −0.7), cg05342682 (β = −5.5%, 95% CI: −9.7, −1.3), cg21856205 (β = −3.1%, 95% CI: −5.5, −0.6), cg01874867 (β = −6.5%, 95% CI: −12.8, −0.3) and cg20119798 (β = −5.3%, 95% CI: −9.8, −0.7). In mid-childhood (n = 149), mean methylation levels of the PON1 DMR were marginally associated with prenatal mercury exposure for males (β = −1.2%, 95% CI: −2.5, 0.1; P = 0.06). However, five individual CpG sites within the PON1 region were associated with maternal RBC-Hg concentrations: cg05342682 (β = −1.2%, 95% CI: −2.5, −0.01), cg19678392 (β = −1.8%, 95% CI: −3.3, −0.3), cg21856205 (β = −0.8%, 95% CI: −1.6, −0.01), cg17330251 (β = −2.1%, 95% CI: −4.1, −0.1) and cg01874867 (β = −2.2%, 95% CI: −4.1, −0.3). Persistence of the association for the male-specific DMR in PON1 and prenatal RBC-Hg concentrations is summarized in Table 2.
Overall CpG-by-CpG analysis in cord blood
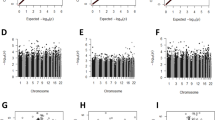
In adjusted robust linear regression models, one CpG was differentially methylated relative to prenatal mercury exposure after using a Bonferroni adjusted level of significance, Fig. 5A. Namely, for every doubling in maternal RBC-Hg concentrations a 0.3% increase in cord blood methylation was observed for cg13340705 annotated to the WW Domain Binding Protein 11 Pseudogene 1 (WBP11P1) located in chromosome 18 (β = 0.3%, 95% CI: 0.2, 0.5; P = 5.3 × 10−7). However, this association did not persist in early (β = −0.1%, 95% CI: −0.5, 0.3; P = 0.57) or mid-childhood (β = 0.03%, 95% CI:−0.2, 0.2; P = 0.81), Table 3.
Sex-specific CpG-by-CpG analysis in cord blood
After stratifying by sex, one CpG in males (cg13416866) annotated to the Torsin Family 4, Member A gene (TOR4A) was hypermethylated relative to maternal RBC-Hg concentrations (β = 1.3%, 95% CI: 0.8, 1.7; P = 3.5 × 10−8), Fig. 5B. The effect estimate for the association between prenatal mercury exposure and methylation levels of this CpG measured during early childhood was similar in direction (β = 0.6%, 95% CI: −0.7, 2.0; P = 0.34) but not significant, likely due to sample size (N = 37). However, the association for this CpG was significant for methylation levels measured in blood at mid-childhood (β = 0.72%, 95% CI: 0.1, 1.4; P = 0.031), Table 3. No sites were found to be differentially methylated in cord blood for females in the CpG-by-CpG analyses, Fig. 5C.
DNA Methylation and Gene Expression
To evaluate the functional relevance of methylation changes, we examined whether DNA methylation of the PON1 DMR and differentially methylated single CpGs were correlated with gene expression in cord blood from an independent birth cohort (Biomarkers of Exposure to Arsenic; N = 38)22. In this independent set of samples we observed a moderate negative correlation between PON1 RNA expression and mean methylation levels of the nine CpGs in the DMR that approached statistical significance (ρ = −0.30, P = 0.06), Fig. 6A. Two CpGs in the PON1 DMR located in the body of the gene and in the north shore region of a CpG island had the strongest association with expression: cg07404485 (ρ = −0.33, P = 0.04) and cg05342682 (ρ = −0.32, P = 0.04), respectively. Five other loci from the DMR reached marginal significance (P < 0.10), Fig. 6B. None of the CpGs found to be associated with prenatal mercury exposure in the CpG-by-CpG analyses correlated with gene expression of cord blood samples, Supplementary Table S1. The genomic tracks of the DMR in PON1 and annotation for the gene expression array used, mRNA, known SNPs and CpGs are shown in Supplementary Fig. S2.
Association between DNA methylation and gene expression of the PON1 gene at the DMR associated with prenatal mercury exposure from an independent birth cohort (N = 38): (A) scatter plot of mean methylation levels of the DMR in PON1 and gene expression and (B) individual correlation coefficients and corresponding P-values for each individual CpG site in the DMR.
DNA Methylation and Child Cognitive Performance
In males, mean methylation levels in cord blood for the PON1 DMR were marginally associated with cognitive PPVT scores measured in early childhood (β = −2.6, 95% CI: −5.6, 0.3; P = 0.07). Within the PON1 DMR, four individual CpGs were significantly associated with cognitive scores measured in early childhood among males. Specifically, an inverse multivariate adjusted association was observed between DNA methylation levels and the PPVT cognitive scores for cg05342682 (β = −3.2, 95% CI: −6.2, −0.2), cg21856205 (β = 4.5, 95% CI: −8.7, −0.1), cg17330251 (β = −1.9, 95% CI: −3.6, −0.1) and cg01874867 (β = −2.0, 95% CI: −3.7, −0.2). No significant associations were observed for individual CpG methylation levels of this region and WRAVMA cognitive test scores in males, Table 4. Although prenatal mercury exposure was not associated with PON1 methylation in females, mean cord blood methylation levels of the PON1 DMR was associated with lower cognitive test scores for the PPVT in girls measured in early childhood (β = −2.6, 95% CI: −4.8, −0.4; P = 0.021) but not WRAVMA scores, Supplementary Table S2. For the CpG-by-CpG analyses methylation levels of cg13340705 in the WBP11P1 gene were marginally associated with WRAVMA cognitive test scores in the entire sample (β = 5.6, 95% CI: −0.6, 11.8; P = 0.07), Supplementary Table S3. Despite previously reported associations between prenatal mercury exposure and cognitive performance within a large subset of the Project Viva cohort8, we did not observe evidence for an association between log2-tranformed RBC-Hg and PPVT cognitive scores (β = 1.2, 95% CI: −1.0, 3.4; P = 0.29) or with WRAVMA scores (β = 0.4, 95% CI: −1.3, 2.0; P = 0.66) in our smaller sample. Therefore, we were unable to estimate the mediated effects of exposure on cognitive performance in this subsample of the cohort.
Finally, we observed similar DNA methylation levels at the PON1 DMR measured in cord blood and in early or mid-childhood (Fig. 7A). Among 21 participants with repeated DNA methylation measurements we observed no significant changes in methylation at the PON1 DMR over time (β = −0.21%; 95% CI: −1.92, 0.78; P = 0.68), Fig. 7B.
Discussion
In this study we observed DNA methylation changes of a genomic region of the PON1 gene relative to prenatal mercury exposure in umbilical cord blood from males but not females, an association that persisted during early and mid-childhood. We also showed that higher DNA methylation levels of the PON1 region are associated with lower cognitive test scores in early childhood for both sexes. Based on publicly available reference datasets we observed that cord blood DNA methylation levels of the PON1 region is associated with decreased PON1 expression in cord blood. In individual CpG analyses, only one CpG of the WBP11P1 gene was associated with prenatal mercury exposure, but this association did not persist in early or mid-childhood. In male-specific site-by-site analyses, a CpG of the TOR4A gene was hypermethylated relative to prenatal mercury exposure and this association persisted in mid-childhood. Taken together, these results suggest that moderate mercury exposure during pregnancy can lead to sex-specific functional epigenetic alterations that persist throughout childhood and are associated with cognitive performance.
The Paraoxonase 1 or arylesterase 1 gene (PON1) belongs to the serum paraoxonase family of enzymes previously shown to provide oxidative protection for high density lipoproteins and hypothesized to guard against atherosclerosis and metabolic syndrome23, 24. PON1 is synthesized in the liver and secreted into the plasma where it can metabolize toxic oxidized lipids25. The main substrates of PON1 are lactones, but promiscuous catalytic activity towards organophosphate triesters, arylesters, cyclic carbamates, glucuronides, oestrogen esters and thiolactones has also been documented26. Genetic polymorphisms in PON1 and enzyme activity have been associated with adverse perinatal health outcomes such as decreased fetal growth, pre-term birth and reductions in head circumference27, 28. Genetic variants as well as low enzymatic activity of PON1 have been shown to contribute to the neurodevelopmental toxicity of organophosphorus (OP) pesticides in animal models and human studies29, 30. PON1 protein has also been localized in brain regions of animal models as well as human tissue and experimentally shown to interact with other proteins in the brain that are essential for brain homeostasis31.
In vitro models and human studies have shown that MeHg exposure exerts an inhibitory effect on PON1 enzyme activity even after controlling for genetic variants including SNP-108 (rs705379), one of the major genetic determinants of PON1 expression32,33,34. The inhibitory mechanism of MeHg on PON1 has been hypothesized to be involved in the development of cardiovascular disease, potentially explaining epidemiological associations with MeHg35. Furthermore, PON1 enzyme activity is lower in neonates but relatively stable in adults exhibiting age-dependent developmental changes through at least 7 years of age. The developmental change in enzyme activity might create a sensitive window for toxic exposures during fetal development25, 36, 37. In fact, hypersensitivity to mercury exposure (acrodynia, or “pink disease”) among infants exposed to mercury has been attributed to genetic variation and subsequent enzyme activity of PON1 38. A large prospective epidemiological study showed that PON1 genotype can modify the association of prenatal MeHg exposure and neurodevelopment, leading to stronger adverse effects in IQ deficits during childhood39. Interestingly, in a study of 896 Inuit adults, blood MeHg concentrations lowered PON1 enzyme activity independent of genetic variation, and there was no evidence of an interaction between MeHg and genetic variants of PON1 32. The lack of gene-environment interaction suggests a third control mechanism involved in the relationship between genetic variation, exposure and subsequent enzyme levels.
Although PON1 genetic variants are better characterized, less is known about epigenetic control mechanisms. In a recent integrative genomic study, a single nucleotide polymorphism in PON1 (SNP-108; rs705379) was shown to be strongly associated with DNA methylation subsequently mediating PON1 enzyme activity in newborns, an association that persisted to nine years of age40. In another study of obese adults, a similar inverse correlation between PON1 methylation and catalytic activity of the enzyme was documented41. This is consistent with our observation in which increasing methylation levels at the PON1 DMR decreased gene expression using data from an independent set of cord blood samples. These results highlight DNA methylation as a control mechanism involved in PON1 expression, underscoring the importance of the male-specific PON1 hypomethylation observed in our study relative to prenatal mercury exposure and subsequent association with cognitive test scores.
Sex-specific differences in DNA methylation for PON1 are unknown for humans. However, in a case-control study of myocardial infarction, female specific hypermethylation of PON1 was observed among cases compared to controls42. Moreover, animal studies have documented male-specific histone modifications along with increased expression of PON1 in male offspring exposed to a high fat diet in utero 43. This is consistent with our data in which male-specific hypomethylation of PON1 was observed for prenatal mercury exposure, potentially leading to an increase in expression. The observed sex-specific epigenetic modification might help explain sex differences observed in mercury’s toxicity. For example, male-specific mercury associations have been documented in epidemiological studies for metabolic syndrome44, ADHD-related behavior, and neurodevelopment45, 46. An experimental study of mice prenatally exposed to mercury demonstrated that males have a higher expression of immune-related genes and miRNA levels compared to females, suggestive of potential sex-specific response during fetal development47.
DNA methylation of the PON1 region could serve as a mediator of genotype and gene expression, explaining age-dependent changes in enzyme activity and modulation of mercury’s neurotoxicity, as shown for OP pesticide exposure. However, our study lacks information on genetic variants to estimate the effect of DNA methylation conditional on genotype. Therefore sex-specific epigenetic differences in the region could partially be driven by genetic variation. Nevertheless, increasing levels of DNA methylation in the PON1 region was associated with a modest decrease in PPVT cognitive scores in both males and females, suggesting that epigenetic control of PON1 could be implicated in cognitive development independent of prenatal mercury exposure or sex. As supported by the observed methylation-expression relationship in the BEAR birth cohort22 and previous studies40, DNA methylation regulates expression and enzymatic activity of PON1 providing a potential mechanism of mercury’s associated neurotoxicity. Furthermore PON1 might also serve as an important target for other toxic effects of mercury exposure.
Although the growing interest in PON1 has been so far focused in cardio-metabolic outcomes, there is evidence that suggests that PON1 genotype and enzyme activity is associated with adult neurological diseases such as Parkinson and Alzheimer48. Furthermore, PON1 genetic variants and enzymatic activity have been linked to autism spectrum disorder in cross-sectional studies49, 50. However, we did not measure DNA methylation in the brain as a target tissue a common limitation of prospective epidemiological studies.
The CpG found to be differentially methylated for the entire sample relative to prenatal mercury exposure is located in the TSS1500 region of the WBP11P1 gene (cg13340705). The WW domain binding protein 11 pseudogene 1 (WBP11P1) has no known function and the association was not persistent in early or mid-childhood suggesting, that this modification is transient at birth. The single CpG (cg13416866) associated with prenatal mercury exposure in males in the Torsin Family 4, Member A gene (TOR4A) persisted into mid childhood but was not associated with cognitive scores. However, these individual loci have not been previously implicated in mercury toxicity, and the evidence for functional or phenotypic association with cognitive performance was limited.
Finally, in the three previous epigenome-wide studies of prenatal mercury exposure no overlaps between differentially methylated regions or CpGs have been documented. One study that included epigenome-wide analysis of placenta samples found that DNA methylation of the EMID2 gene was associated with prenatal mercury exposure and neonate behavior. Interestingly both the EMID2 and PON1 gene are located in a region of chromosome 7 (q21–q31), experimentally shown to be sensitive to genomic imprinting using human-mouse monochromosomal hybrids51. Additionally, this region in PON1 has been shown to have parent of origin specific methylation involved in the etiology of Silver-Russell syndrome, but PON1 remains a disputed imprinted gene in humans52. Future studies should evaluate if prenatal mercury exposure targets other imprinted genes from this region. Heterogeneity of the DNA methylation targets across studies suggests that mercury’s associated epigenetic disruption might be time-sensitive during fetal development, emphasizing the importance of collecting both sensitive and time-specific biomarkers of exposure. Furthermore, mercury exposure levels in our cohort were relatively low but comparable to the general US population53. Therefore our results might not be generalizable to populations exposed to higher levels of mercury.
Our study has several limitations. First, in the absence of genotype information it is not possible to determine if genetic variants are responsible for the observed methylation differences. However, it has been shown that MeHg exposure inhibits PON1 activity even after controlling for the major known genetic variants, suggesting a dominant epigenetic control mechanism for the enzyme. While we observed associations between DNA methylation and gene expression these associations may not be generalizable to our cohort. Another important limitation of our study is the use of adult reference DNA methylation data to impute white blood cell distribution, as adult blood might not accurately capture leukocyte distribution in childhood. However, we used a cord blood reference panel to estimate cell proportions at birth, including nucleated red blood cells, and the persistence of epigenetic modifications were robust to white blood cell composition adjustment using different panels for cord blood and child blood. We also lack mercury exposure information during early and mid-childhood or any pesticide exposure information and therefore the relative contribution of postnatal exposures cannot be determined. The study was observational in nature and confounding cannot be ruled out.
The present study has many strengths that include the use of second trimester maternal red-blood cell mercury concentrations, an unbiased biomarker with an estimated half-life of 72 days54 and over 70% of the red-blood cell mercury is estimated to be methylmercury55. It is expected that this biomarker captured prenatal exposures occurring during critical windows of embryogenesis and fetal development. Although our sample size is moderate, this is the largest epigenome-wide study of prenatal mercury exposure conducted to date and the only one to prospectively test for persistence of epigenetic modifications at two time points during childhood. Our moderate sample size enabled us to evaluate sex-specific differences. Furthermore, this cohort has detailed anthropomorphic and demographic information that allowed us to control for many potential confounders and test for associations with cognitive test scores during early childhood. Detailed information on maternal diet also allowed us to control for fish intake, previously shown to be beneficial for children’s cognitive development, minimizing the possibility for negative confounding8. The analyses of regional and individual CpG sites methylation variability is also an important strength of the present study. Lastly, the potential for functional changes in DNA methylation with gene expression was evaluated in a separate set of cord blood samples further supporting our findings.
This study adds to the growing body of evidence that suggests that PON1 serves as a potential target of toxic environmental exposures particularly during fetal development and childhood. Furthermore, DNA methylation of PON1 could modulate the association between prenatal mercury exposure, cognitive development and other health outcomes in children. Epigenetic modifications leading to functional genomic changes could help explain heterogeneous findings observed for prenatal mercury exposure and cognitive development in different populations. This will also provide tools to design future targeted public health interventions.
Methods
Study Population
Mothers and children were participants in Project Viva, a prospective pre-birth cohort study conducted in Massachusetts, USA56. This cohort was recruited between 1999 and 2002 during their first prenatal visit at Atrius Harvard Vanguard Medical Associates, a large multispecialty group practice. Mothers provided written informed consent at study recruitment and at every follow-up visit. All study protocols were reviewed and carried out in accordance with guidelines approved by the human subjects committee of Harvard Pilgrim Health Care. Eligibility criteria included fluency in English, gestational age less than 22 weeks at the first prenatal visit, and singleton pregnancy. Additional details of the cohort have been published elsewhere56. Of the total 2,128 mother-infants pairs in the cohort, we measured prenatal mercury exposure from 1019. Of these, we included 321 with available measurements of cord blood DNA methylation. Persistence of the epigenetic modifications were evaluated in 75 children with available whole blood DNA methylation measurements from early childhood (2.9 to 4.9 years) and 291 children with blood samples from mid-childhood (6.7 to 10.5 years) as well as prenatal mercury concentrations. Early and mid-childhood participants were not restricted to participants with cord blood samples and therefore are not mutually inclusive. Of the 321 infants eligible for analyses with cord blood samples, 70 were included for early childhood analyses and 160 in the mid-childhood analyses. The number of participants sampled in both early and mid-childhood was 43.
Red Blood Cell Mercury (RBC-Hg) and Fish Intake
Sample collection and exposure assessment for prenatal mercury exposure in this cohort has been previously described in detail8. Briefly, at the second trimester visit we obtained a maternal blood sample that was centrifuged to separate plasma from erythrocytes. Erythrocyte aliquots were stored at −70 °C and analyzed for total mercury by using the Direct Mercury Analyzer 80 (Milestone Inc., Monroe, Connecticut). Results are reported as mercury content in the original red blood cell sample. The detection limit was 0.5 ng/g of sample, and the percentage recovery for standards ranged from 90 to 110%. At mid-pregnancy, participants also completed a semi-quantitative food frequency questionnaire that we previously calibrated against erythrocyte levels of elongated n-3 fatty acids57. Participants self-reported on the consumption of fish with four questions: “canned tuna fish (3–4 oz.)”; “shrimp, lobster, scallops, clams (1 serving)”; “dark meat fish, e.g. mackerel, salmon, sardines, bluefish, swordfish (3–5 oz.)”; and “other fish, e.g. cod, haddock, halibut (3–5 oz.)”, during the preceding 3 months. Six frequency response options ranged from “never/less than 1 per month” to “1 or more servings per day.” We combined responses to estimate average total fish intake in servings per week.
DNA Extraction and Sample Collection
Trained medical personnel obtained umbilical cord blood samples immediately upon delivery storing them in a dedicated refrigerator at 4 °C and transported to a central location within 24 hours of sample collection. Similarly, whole blood samples collected during early, and mid-childhood were stored at 4 °C and transported to the central storage location for sample processing. Subsequently, trained laboratory staff processed the samples on the same day of arrival, and DNA was extracted using the Qiagen Puregene Kit (Valencia, CA). Aliquots were then stored at −80 °C until analysis.
DNA methylation Assessment and Quality Control
Buffy coat DNA was sodium bisulfite converted using the EZ DNA Methylation-Gold Kit (Zymo Research, Irvine, CA). Samples were allocated to plates using a two-stage algorithm by randomizing 12 samples to each chip and then randomly assigning eight chips to each of the 15 plates used to ensure balance by sex across chips and plates. Samples were shipped to Illumina Inc., and analyzed using the Infinium Human Methylation450 BeadChip (Illumina, San Diego, CA) following standard manufacturer’s protocols. The Human Methylation450 BeadChip measures DNA methylation at >485,000 CpG sites simultaneously at a single nucleotide resolution, covering 99% of the RefSeq genes.
We processed raw methylation image files using the minfi package in R58. Samples were excluded as potentially miss-labelled if they were mismatches on sex (n = 6), genotype (n = 6) or were deemed to be low in quality (n = 12). Technical replicates were also excluded from the analysis (n = 40). Correlation coefficients for individual probes among all technical replicates ranged from 0.98 to 1. We excluded individual probes if they had non-significant detection P-values (P > 0.05) for more than 1% of the samples. Additionally, non-CpG probes (i.e. rs and ch), probes in X and Y chromosomes, SNP-associated probes at either the single base extension or within the target region were removed for SNPs that have a minor-allele frequency of >5%. Previously identified non-specific and cross-reactive probes within the array along with polymorphic CpG loci were also excluded from the analysis59. After quality control on the probes, the total number of autosomal CpGs left for analysis was 384,349 loci for all the cord blood samples. Background correction and dye-bias equalization was performed via the normal-exponential out-of-band (noob) correction method60. Finally, a β-mixture quantile intra sample normalization procedure (BMIQ) was applied to the resulting data to reduce the potential bias that can arise from type2 probes59. For each CpG site, methylation is reported as average β-value = M/(M + U + ε), where M and U represent the average fluorescence intensity from the probe corresponding to the methylated and unmethylated target CpG and ε = 100 is a small quantity to protect against division by zero. Thus, the average β-value is an interval scaled quantity between zero and one interpreted as the fraction of DNA molecules whose target CpG is methylated.
We used ComBat61 to correct for Batch effects from plate and other potential sources of technical variability in methylation measurements. We visually inspected the effectiveness of adjustment for batch using the four main principal components before and after batch adjustment. Strip plots of control probes were visually examined for bisulfite conversion and specificity. Density plots for the β-values were examined across samples at each normalization step. Methylation values on the β-scale were logit transform to M-values as previously described to be more appropriate for differential analysis of DNA methylation62. All tables and results are presented on the β-value scale to ease interpretability.
Cognitive Outcomes in Early Childhood
At the early childhood visit, trained research personnel administered the Peabody Picture Vocabulary Test (PPVT) and the Wide Range Assessment of Visual Motor Abilities (WRAVMA) cognitive tests. The PPVT evaluates receptive vocabulary for children age 2 or older based on a national reference sample and it is strongly correlated (r > 0.90) with verbal and full-scale intelligence quotient of the Wechsler Intelligence Scale for Children-III63. The Wide Range Assessment of Visual Motor Abilities (WRAVMA) evaluates domains of visual motor development and is moderately correlated with intelligence quotient (r~0.60)64.
Statistical Analysis
We calculated means and standard deviations (SD) or sample size and percentage for all covariates to describe the study population during the three time points: birth, early childhood (2.9 to 4.9 years) and mid-childhood (6.7 to 10.5 years). White blood cell composition was estimated from DNA methylation measurements using the Houseman projection method65 from isolated cell types. To estimate cell types composition in cord blood we used a reference panel of nucleated cells isolated from cord blood (leukocytes and nucleated red blood cells)66 and an adult leukocyte reference panel for blood samples collected in early or mid-childhood as implemented in minfi 58, 67. We conducted a principal component analysis in the resulting 384,349 CpGs across all cord blood samples and used univariate linear regression models to explore associations among each of the top 30-principal components with phenotypic and technical covariates using the EnMix Bioconductor package of R68. All analyses were carried out using the R statistical package, version 3.2.3 (www.r-project.org/).
Differentially Methylated Regional Analyses
We examined the association of prenatal mercury exposure with differentially methylated regions (DMRs) in cord blood using the R Bioconductor package DMRcate69. This regional analysis method was chosen over other available methods because it has an agnostic approach to both genomic annotation and direction of effect estimates. Briefly, this package first fits individual linear regression models using limma for each CpG and subsequently applies a Gaussian kernel smoothing function to test statistics grouping significant probes based on a maximum distance of λ = 1,000 base pairs. Significance testing among DMRs are adjusted for multiple comparisons using an FDR < 0.05. Regional analyses were performed in cord blood for all of the samples and also stratified by sex while adjusting for child gestational age at delivery, sex (if not stratified), and estimated nucleated cell types in cord blood (CD8+, CD4+, Natural Killer cells, Monocytes, B-cells and nucleated red blood cells). As well as maternal age, race/ethnicity, mean weekly fish intake during pregnancy, pre-pregnancy BMI, smoking during pregnancy, parity and college education.
Among significant DMRs found relative to log2-transformed mercury exposure we used robust linear models to estimate the association of each CpG in the region as well as mean methylation levels of all CpG sites in the DMR relative to maternal RBC-Hg concentrations adjusting for potential confounders. Persistence of the epigenetic changes was evaluated in early and mid-childhood blood DNA using similar robust linear models. Multivariate linear models were adjusted for the same covariates as the cord blood models with the exceptions that child race was used instead of maternal race, white blood cell composition was estimated from whole blood DNA methylation using an adult reference panel, and models were further adjusted by age of the child in days at the time of the blood draw. Associations were considered to be persistent with P < 0.05.
CpG-by-CpG Analyses
We evaluated methylation differences at individual CpG sites in cord blood relative to maternal RBC-Hg concentrations using robust linear regression with heteroskedasticity-consistent estimators to model the methylation levels of each individual CpG on the M-value scale as the dependent variable and log2-transformed prenatal mercury exposure as the main predictor while adjusting for the same covariates as in the regional analyses. CpG-by-CpG analyses were further stratified by infant sex to evaluate sex-specific epigenetic disruption at individual sites. The genomic inflation factor (λ) for the overall analysis was 0.99 indicating of no major systemic biases. Statistical significance for the CpG-by-CpG analyses was evaluated using a Bonferroni adjusted level of significance (P < 1.3 × 10−7). Although the epigenome-wide analysis was performed on the M-value scale, all estimates are reported as percent change in methylation along with the corresponding P-values from robust linear regression models performed on the β-value scale, for ease of interpretability.
We carried forward individual loci with a P < 1.3 × 10−7 in cord blood analyses to subsequent analyses for blood samples collected during early and mid-childhood to evaluate the persistence of associations using multivariate robust linear regression models adjusting for covariates. We considered P < 0.05 as indicating the persistence of epigenetic alterations in early or mid-childhood for the CpG-by-CpG analyses.
DNA Methylation and Gene Expression
To evaluate if the epigenetic alterations observed relative to prenatal mercury exposure are related to functional genomic changes in cord blood (i.e. expression), we used two publicly available datasets from an independent birth cohort found in the Gene Expression Omnibus (GEO) repository. Specifically, we identified DNA methylation data from 38 cord blood samples that used the Illumina Human Methylation450 array (GSE62924) and paired them with gene expression profiles from the same subjects (GSE48355) measured using the Affymetrix Human Gene 2.0 ST array. The study design, methods, sample collection and processing for this birth cohort have been previously described22.
We estimated correlation coefficients between gene expression and DNA methylation at CpGs within the DMR and at individual sites observed to be differentially methylated in cord blood relative to prenatal mercury exposure in our data. For the DMR found to be associated with prenatal mercury exposure, correlation coefficients between expression and mean methylation levels of the region as well as at individual CpGs within the region were tested. Correlation coefficients along with 95% Confidence Intervals (95% CIs) were used to estimate the association.
Cord Blood DNA Methylation and Cognitive Performance
Linear regression models were used to estimate the association between differentially methylated DMRs or CpGs found in cord blood and cognitive test scores (PPVT and WRAVMA). Models were adjusted for maternal education at study enrollment, maternal PPVT scores, self-reported alcohol use during pregnancy, fetal growth (sex-specific z-score of birth weight/gestational age), mean weekly fish intake during pregnancy, child age in days at the time of testing, sex (if not stratified), child race, parity and maternal smoking during pregnancy. Estimates are reported for every 10% increase in methylation levels along with 95% CIs.
References
Swain, E. B. et al. Socioeconomic consequences of mercury use and pollution. AMBIO: A Journal of the Human Environment 36, 45–61 (2007).
Lamborg, C. H. et al. A global ocean inventory of anthropogenic mercury based on water column measurements. Nature 512, 65–68 (2014).
Goodrich, J. M., Basu, N., Franzblau, A. & Dolinoy, D. C. Mercury biomarkers and DNA methylation among Michigan dental professionals. Environmental and molecular mutagenesis 54, 195–203 (2013).
Stern, A. H. & Smith, A. E. An assessment of the cord blood: maternal blood methylmercury ratio: implications for risk assessment. Environmental health perspectives 111, 1465 (2003).
Harada, M. Minamata disease: methylmercury poisoning in Japan caused by environmental pollution. CRC Critical Reviews in Toxicology 25, 1–24 (1995).
Grandjean, P. et al. Cognitive deficit in 7-year-old children with prenatal exposure to methylmercury. Neurotoxicology and teratology 19, 417–428 (1997).
Oken, E. et al. Maternal fish consumption, hair mercury, and infant cognition in a US cohort. Environmental health perspectives. 1376–1380 (2005).
Oken, E. et al. Maternal fish intake during pregnancy, blood mercury levels, and child cognition at age 3 years in a US cohort. American Journal of Epidemiology 167, 1171–1181 (2008).
Freire, C. et al. Hair mercury levels, fish consumption, and cognitive development in preschool children from Granada, Spain. Environmental research 110, 96–104 (2010).
Boucher, O. et al. Domain-specific effects of prenatal exposure to PCBs, mercury, and lead on infant cognition: results from the Environmental Contaminants and Child Development Study in Nunavik. Environmental health perspectives 122, 310 (2014).
Karagas, M. et al. Evidence on the human health effects of low level methylmercury exposure. Environmental health perspectives 120, 799–806 (2012).
Perera, F. & Herbstman, J. Prenatal environmental exposures, epigenetics, and disease. Reproductive toxicology 31, 363–373 (2011).
Vaiserman, A. Epidemiologic evidence for association between adverse environmental exposures in early life and epigenetic variation: a potential link to disease susceptibility? Clinical epigenetics 7(1), 96 (2015).
Acevedo, N. et al. Age-associated DNA methylation changes in immune genes, histone modifiers and chromatin remodeling factors within 5 years after birth in human blood leukocytes. Clinical epigenetics 7(1), 34 (2015).
Cardenas, A. et al. Differential DNA methylation in umbilical cord blood of infants exposed to mercury and arsenic in utero. Epigenetics 10, 508–515 (2015).
Bakulski, K. M. et al. Prenatal mercury concentration is associated with changes in DNA methylation at TCEANC2 in newborns. International journal of epidemiology 44, 1249–1262 (2015).
Maccani, J. Z. et al. Placental DNA methylation related to both infant toenail mercury and adverse neurobehavioral outcomes. Environmental health perspectives 123, 723 (2015).
Bale, T. L. Epigenetic and transgenerational reprogramming of brain development. Nature Reviews Neuroscience 16, 332–344 (2015).
van Dongen, J. et al. Genetic and environmental influences interact with age and sex in shaping the human methylome. Nature Communications 7 (2016).
Vahter, M., Åkesson, A., Lidén, C., Ceccatelli, S. & Berglund, M. Gender differences in the disposition and toxicity of metals. Environmental research 104, 85–95 (2007).
Ruszkiewicz, J. A., Bowman, A. B., Farina, M., Rocha, J. B. & Aschner, M. Sex-and structure-specific differences in antioxidant responses to methylmercury during early development. Neurotoxicology 56, 118–126 (2016).
Rojas, D. et al. Prenatal arsenic exposure and the epigenome: identifying sites of 5-methylcytosine alterations that predict functional changes in gene expression in newborn cord blood and subsequent birth outcomes. Toxicological Sciences 143, 97–106 (2015).
Huen, K., Harley, K., Beckman, K., Eskenazi, B. & Holland, N. Associations of PON1 and genetic ancestry with obesity in early childhood. PloS one 8, e62565 (2013).
Bhattacharyya, T. et al. Relationship of paraoxonase 1 (PON1) gene polymorphisms and functional activity with systemic oxidative stress and cardiovascular risk. Jama 299, 1265–1276 (2008).
Costa, L. G., Vitalone, A., Cole, T. B. & Furlong, C. E. Modulation of paraoxonase (PON1) activity. Biochemical pharmacology 69, 541–550 (2005).
Mackness, M. & Mackness, B. Human paraoxonase-1 (PON1): Gene structure and expression, promiscuous activities and multiple physiological roles. Gene 567, 12–21 (2015).
Berkowitz, G. S. et al. In utero pesticide exposure, maternal paraoxonase activity, and head circumference. Environmental health perspectives 112, 388 (2004).
Infante-Rivard, C. Genetic association between single nucleotide polymorphisms in the paraoxonase 1 (PON1) gene and small-for-gestational-age birth in related and unrelated subjects. American journal of epidemiology 171, 999–1006 (2010).
Costa, L. G., Giordano, G., Cole, T. B., Marsillach, J. & Furlong, C. E. Paraoxonase 1 (PON1) as a genetic determinant of susceptibility to organophosphate toxicity. Toxicology 307, 115–122 (2013).
Eskenazi, B. et al. PON1 and neurodevelopment in children from the CHAMACOS study exposed to organophosphate pesticides in utero. Environmental health perspectives 118, 1775 (2010).
Suszyńska-Zajczyk, J., Łuczak, M., Marczak, Ł. & Jakubowski, H. Inactivation of the paraoxonase 1 gene affects the expression of mouse brain proteins involved in neurodegeneration. Journal of Alzheimer’s Disease 42, 247–260 (2014).
Ayotte, P. et al. Relation between methylmercury exposure and plasma paraoxonase activity in Inuit adults from Nunavik. Environmental health perspectives 119, 1077 (2011).
Brophy, V. H. et al. Effects of 5′ regulatory-region polymorphisms on paraoxonase-gene (PON1) expression. The American journal of human genetics 68, 1428–1436 (2001).
Drescher, O. et al. Methylmercury exposure, PON1 gene variants and serum paraoxonase activity in Eastern James Bay Cree adults. Journal of Exposure Science and Environmental Epidemiology 24, 608–614 (2014).
Ginsberg, G., Sonawane, B., Nath, R. & Lewandowski, P. Methylmercury-induced inhibition of paraoxonase-1 (PON1)—Implications for cardiovascular risk. Journal of Toxicology and Environmental Health, Part A 77, 1004–1023 (2014).
Huen, K. et al. Developmental changes in PON1 enzyme activity in young children and effects of PON1 polymorphisms. Environmental health perspectives 117, 1632 (2009).
Chen, J., Kumar, M., Chan, W., Berkowitz, G. & Wetmur, J. G. Increased influence of genetic variation on PON1 activity in neonates. Environmental Health Perspectives 111, 1403 (2003).
Austin, D. W. et al. Genetic variation associated with hypersensitivity to mercury. Toxicology international 21, 236 (2014).
Julvez, J. et al. Prenatal methylmercury exposure and genetic predisposition to cognitive deficit at age 8 years. Epidemiology 24, 643–650 (2013).
Huen, K., Yousefi, P., Street, K., Eskenazi, B. & Holland, N. PON1 as a model for integration of genetic, epigenetic, and expression data on candidate susceptibility genes. Environmental Epigenetics 1, dvv003 (2015).
de la Iglesia, R., Mansego, M. L., Sánchez-Muniz, F. J., Zulet, M. A. & Martinez, J. A. Arylesterase activity is associated with antioxidant intake and paraoxonase-1 (pon1) gene methylation in metabolic syndrome patients following an energy restricted diet. EXCLI journal 13, 416 (2014).
Fiorito, G. et al. B-vitamins intake, DNA-methylation of one carbon metabolism and homocysteine pathway genes and myocardial infarction risk: the EPICOR study. Nutrition, Metabolism and Cardiovascular Diseases 24, 483–488 (2014).
Strakovsky, R. S., Zhang, X., Zhou, D. & Pan, Y.-X. The regulation of hepatic Pon1 by a maternal high-fat diet is gender specific and may occur through promoter histone modifications in neonatal rats. The Journal of nutritional biochemistry 25, 170–176 (2014).
Chung, J.-Y., Seo, M.-S., Shim, J.-Y. & Lee, Y.-J. Sex differences in the relationship between blood mercury concentration and metabolic syndrome risk. Journal of endocrinological investigation 38, 65–71 (2015).
Sagiv, S. K., Thurston, S. W., Bellinger, D. C., Amarasiriwardena, C. & Korrick, S. A. Prenatal exposure to mercury and fish consumption during pregnancy and attention-deficit/hyperactivity disorder–related behavior in children. Archives of pediatrics & adolescent medicine 166, 1123–1131 (2012).
Gao, Y. et al. Prenatal exposure to mercury and neurobehavioral development of neonates in Zhoushan City, China. Environmental research 105, 390–399 (2007).
Penta, K. L., Altomare, D., Shirley, D. L. & Nyland, J. F. Female immune system is protected from effects of prenatal exposure to mercury. (2015).
Androutsopoulos, V. P., Kanavouras, K. & Tsatsakis, A. M. Role of paraoxonase 1 (PON1) in organophosphate metabolism: implications in neurodegenerative diseases. Toxicology and applied pharmacology 256, 418–424 (2011).
D’amelio, M. et al. Paraoxonase gene variants are associated with autism in North America, but not in Italy: possible regional specificity in gene–environment interactions. Molecular psychiatry 10, 1006–1016 (2005).
Paşca, S. P. et al. Paraoxonase 1 activities and polymorphisms in autism spectrum disorders. Journal of cellular and molecular medicine 14, 600–607 (2010).
Okita, C. et al. A new imprinted cluster on the human chromosome 7q21-q31, identified by human-mouse monochromosomal hybrids. Genomics 81, 556–559 (2003).
Hannula-Jouppi, K. et al. Differentially methylated regions in maternal and paternal uniparental disomy for chromosome 7. epigenetics 9, 351–365 (2014).
Oken, E. et al . Maternal prenatal fish consumption and cognition in mid childhood: Mercury, fatty acids, and selenium. Neurotoxicology and Teratology [Epub ahead of print] (2016).
Mahaffey, K. R., Clickner, R. P. & Bodurow, C. C. Blood organic mercury and dietary mercury intake: National Health and Nutrition Examination Survey, 1999 and 2000. Environmental health perspectives 112, 562 (2004).
Mortensen, M. E., Caudill, S. P., Caldwell, K. L., Ward, C. D. & Jones, R. L. Total and methyl mercury in whole blood measured for the first time in the US population: NHANES 2011–2012. Environmental research 134, 257–264 (2014).
Oken, E. et al. Cohort profile: project viva. International journal of epidemiology 44, 37–48 (2014).
Fawzi, W. W., Rifas-Shiman, S. L., Rich-Edwards, J. W., Willett, W. C. & Gillman, M. W. Calibration of a semi-quantitative food frequency questionnaire in early pregnancy. Annals of epidemiology 14, 754–762 (2004).
Aryee, M. J. et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 30, 1363–1369, doi:10.1093/bioinformatics/btu049 (2014).
Teschendorff, A. E. et al. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics 29, 189–196 (2013).
Triche, T. J. Jr., Weisenberger, D. J., Van Den Berg, D., Laird, P. W. & Siegmund, K. D. Low-level processing of Illumina Infinium DNA Methylation BeadArrays. Nucleic acids research 41, e90, doi:10.1093/nar/gkt090 (2013).
Johnson, W. E., Li, C. & Rabinovic, A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8, 118–127 (2007).
Du, P. et al. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC bioinformatics 11, 587 (2010).
Dunn, L. M. & Dunn, L. M. Examiner’s manual for the Peabody Picture Vocabulary Test. 3rd ed. Circle Pines, MN: American Guidance Service (1997).
Adams, W. & Sheslow, D. Wide range assessment of visual motor abilities. Wilmington, DE: Wide Range, Inc (1995).
Houseman, E. A. et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC bioinformatics 13, 1 (2012).
Bakulski, K. M. et al. DNA methylation of cord blood cell types: Applications for mixed cell birth studies. Epigenetics 00–00 (2016).
Reinius, L. E. et al. Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PloS one 7, e41361 (2012).
Xu, Z., Niu, L., Li, L. & Taylor, J. A. ENmix: a novel background correction method for Illumina Human Methylation450 BeadChip. Nucleic acids research. gkv907 (2015).
Peters, T. J. et al. De novo identification of differentially methylated regions in the human genome. Epigenetics & chromatin 8, 1 (2015).
Acknowledgements
This work was supported by grants from the National Institutes of Health (R01 NR013945, R01 ES021357, R37 HD034568, K24 HD069408, R01 ES016314 and R01 HL 111108).
Author information
Authors and Affiliations
Contributions
Andres Cardenas is the lead author of the study and carried out all genome-wide analyses with the guidance and supervision of Andrea A. Baccarelli. Sheryl L. Rifas-Shiman, Golareh Agha, Marie-France Hivert, Augusto A. Litonjua, Dawn Demeo, Xihong Lin, Chitra J. Amarasiriwardena, Emily Oken and Mathew W. Gillman helped conceptualized the project, provided helped with different aspects of the analysis and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Cardenas, A., Rifas-Shiman, S.L., Agha, G. et al. Persistent DNA methylation changes associated with prenatal mercury exposure and cognitive performance during childhood. Sci Rep 7, 288 (2017). https://doi.org/10.1038/s41598-017-00384-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-00384-5
This article is cited by
-
Sex difference of pre- and post-natal exposure to six developmental neurotoxicants on intellectual abilities: a systematic review and meta-analysis of human studies
Environmental Health (2023)
-
Prenatal Exposure to Potentially Toxic Metals and Their Effects on Genetic Material in Offspring: a Systematic Review
Biological Trace Element Research (2023)
-
Epigenetics as a Biomarker for Early-Life Environmental Exposure
Current Environmental Health Reports (2022)
-
A pilot study that provides evidence of epigenetic changes among mother–child pairs living proximal to mining in the US
Environmental Geochemistry and Health (2022)
-
Effect of methylmercury on fetal neurobehavioral development: an overview of the possible mechanisms of toxicity and the neuroprotective effect of phytochemicals
Archives of Toxicology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.