Abstract
Understanding the lower limb kinematic, kinetic, and electromyography (EMG) data interrelation in controlled speeds is challenging for fully assessing human locomotion conditions. This paper provides a complete dataset with the above-mentioned raw and processed data simultaneously recorded for sixteen healthy participants walking on a 10 meter-flat surface at seven controlled speeds (1.0, 1.5, 2.0, 2.5, 3.0, 3.5, and 4.0 km/h). The raw data include 3D joint trajectories of 24 retro-reflective markers, ground reaction forces (GRF), force plate moments, center of pressures, and EMG signals from Tibialis Anterior, Gastrocnemius Lateralis, Biceps Femoris, and Vastus Lateralis. The processed data present gait cycle-normalized data including filtered EMG signals and their envelope, 3D GRF, joint angles, and torques. This study details the experimental setup and presents a brief validation of the data quality. The presented dataset may contribute to (i) validate and enhance human biomechanical gait models, and (ii) serve as a reference trajectory for personalized control of robotic assistive devices, aiming an adequate assistance level adjusted to the gait speed and user’s anthropometry.
Measurement(s) | walking • Lower Extremity |
Technology Type(s) | motion capture • electromyography • force platform |
Factor Type(s) | sex • age • height • mass • leg length • foot length |
Sample Characteristic - Organism | Homo sapiens |
Machine-accessible metadata file describing the reported data: https://doi.org/10.6084/m9.figshare.13169348
Similar content being viewed by others
Background & Summary
Walking appears to be the most performed human motor task1. However, healthy human gait may be compromised by neurological diseases (such as stroke and spinal cord injury)2,3. Robotic assistive devices (namely, active orthosis and exoskeletons) have been recommended to foster repetitive gait rehabilitation sessions for patients with locomotor impairments that lost the ability to walk. Typically, robotic devices endow control architectures that require reference trajectories (e.g., joint angles, torques, or muscle activations) to determine the amount of needed assistance or to define a target walking pattern4. These reference trajectories are commonly obtained from public walking datasets from healthy users’ self-selected speeds.
Moreover, control architectures may include objective assessment tools of the patient’s disability level as a fundamental point for developing personalized rehabilitation strategies oriented to the patient’s locomotion condition5. Kinematic, kinetic, and electromyography (EMG) data are commonly used to describe the human locomotion condition6. Typically, clinical studies distinguish pathological and healthy gait by using these walking data collected at healthy users’ self-selected speeds, available in the literature7,8,9.
Nonetheless, participants with lower limb impairments present slower speeds when compared to the healthy gait at self-selected speeds10. Therefore, the validity of applying reference trajectories in robotic devices and the pathological vs healthy gait pattern distinction may be compromised due to the potential bias that can be introduced by the speed differences in the gait dynamics4,11,12,13,14. Additionally, the comprehension of the body mass and height effects on lower limb biomechanical parameters is fundamental to determine changes in the gait pattern4,13,15,16,17. Thus, the provision of multimodal walking datasets oriented to the anthropometry and walking speed conditions of each participant has been highlighted.
Although the high number of performed gait studies, there are still a few available multimodal walking datasets. Furthermore, most of them present the following shortcomings. First, only provide the average and the standard deviation of the data for a single gait cycle for all the participants that participated in each study, not providing the raw data12. In this manner, it is not possible to access the walking data considering the anthropometry and speed of each participant. Additionally, some available datasets only foster data for healthy self-selected speeds5,12,18,19,20. On the other hand, some datasets do not provide EMG data, not being suitable for applications that depend on EMG information19,20,21. Despite being suitable for several applications, the usefulness of these datasets can be restricted.
In this study, we present a multimodal walking dataset containing EMG data from four muscles (Tibialis Anterior (TA), Gastrocnemius Lateralis (GAL), Biceps Femoris (BF), and Vastus Lateralis (VL)), 3D ground reaction forces (GRFs), Center of Pressures (CoPs), and joint angles and torques of the lower limb joints (hip, knee, and ankle) along with the pelvis segment. The data were collected from sixteen healthy participants performing walking trials at seven controlled speeds (1.0, 1.5, 2.0, 2.5, 3.0, 3.5, and 4.0 km/h, corresponding to 0.28, 0.42, 0.56, 0.69, 0.83, 0.97, and 1.1 m/s, respectively) on a flat surface. The gait speed selection was based on the (i) preferred speed of the most incident population with gait impairments and (ii) comfortable speed of healthy participants10,22. Since the joint biomechanics during the walking motion depend on gender, body height, and mass, the proposed dataset tackles an equal gender distribution with a wide range of body heights (ranging from 1.51 to 1.83 m) and mass (ranging from 52.0 to 83.7 kg)23,24. Based on the best knowledge of the authors, this is the first study that presents a database that gathers anthropometric information, synchronized EMG, 3D GRF, CoPs, and angles and torques data of the lower limb joints along with the pelvis segment under controlled speeds.
The proposed multimodal walking dataset may contribute to (i) the development of biomechanical gait models scaled to variable anthropometric parameters and speeds; (ii) assess the human locomotion conditions based on lower limb kinematic, kinetic, and EMG interrelation at controlled speeds; (iii) perform a biomechanical analysis of gait and muscle activation patterns during speed variation; and (iv) serve as reference trajectories for robotic devices, aiming an adequate assistance level adjusted to the speed and user’s anthropometry.
Methods
Participants
Healthy participants were approached through a mailing list across the University of Minho community describing the study goal, protocol, and duration. The participants were formally recruited and selected according to a set of inclusion criteria, as follows: (i) healthy locomotion; (ii) more than 18 years old; (iii) body mass ranging from 45 to 90 kg; and, (iv) body height ranging from 1.50 to 1.90 m. The participants disclaimed a history of injuries or musculoskeletal dysfunctions. These criteria were followed to produce a dataset with a stratified anthropometric distribution while considering possible gender biomechanical differences23,24,25. In this connection, sixteen healthy participants (8 males and 8 females) were involved in the study. They present a mean age of 23.8 ± 2.02 years, a mean body mass of 67.5 ± 10.8 kg, and a mean body height of 1.69 ± 0.108 m, as presented in Table 1. Before undergoing the experiments, all participants provided written and informed consent, according to the ethical conduct defined by the University of Minho Ethics Committee that follows the standard set by the declaration of Helsinki and the Oviedo Convention (CEICVS 006/2020). Moreover, participants also provided written consent to publish identifiable images.
Instrumentation & data collection
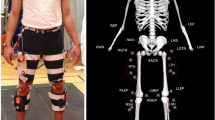
Data were collected at Porto Biomechanics Laboratory (LABIOMEP). The participants were instrumented with a surface EMG system and lower body motion-capture system, as illustrated in Fig. 1. The participants were asked to wear tight shorts and a standard type of tennis shoes that properly fits the user’s feet.
EMG sensors attached to the participant’s skin: (A) - VL, (B) - TA, (C) - BF, and (D) - GAL. Retro-reflective markers: 1 - Anterior Superior Iliac Spine (ASIS), 2 - Trochanter (TROC), 3 - Thigh (TH), 4 - Lateral Knee (LK), 5 - Medial Knee (MK), 6 - Shank (SK), 7 - Lateral Ankle (LA), 8 - Medial Ankle (MA), 9 - Foot Metatarsal 5 (FM5), 10 - Foot Metatarsal 1 (FM1), 11 - Foot Metatarsal 2 (FM2), 12 - Posterior Superior Iliac Spine (PSOS). Data acquisition scenario, where E and F represent the beginning and the end position of the recording, respectively.
A surface 8-channel wireless EMG system (Trigno – Delsys, Massachusetts, United States of America), acquiring at 2000 Hz, provided the EMG signals from four lower limb muscles, namely, TA, GAL, BF, and VL of both lower limbs (as presented in Fig. 1). Prior to the EMG sensors’ placement, the skin above each muscle was gently cleaned with alcohol wipes to remove skin and dirt oils. Once cleaned, the eight EMG electrodes (Trigno – Delsys, Massachusetts, bipolar configuration with an interelectrode distance of 10 mm) were attached to the skin with double-sided adhesive tape, following the SENIAM recommendations26 (Fig. 1). The quality of all EMG signals was checked, and the location of the sensors was adjusted when required. To avoid EMG sensor disconnection from the human skin during the data acquisition, each sensor was fixed with a strap, as presented in Fig. 1, without introducing erroneous measures.
3D kinematic motion information from the three lower limb joints (hip, knee, and ankle) and pelvis segment was measured at 200 Hz using a motion-capture system with 12 cameras (Oqus, Qualisys – Motion-Capture System, Göteborg, Sweden). Each participant was equipped with the Newington-Helen Hayes marker-set, integrating four more retro-reflective markers placed on the trochanter, medial tuberosity of the femur, medial malleolus, and first metatarsal head to support the calibration process27. Overall, 12 pairs of retro-reflective markers were used, as depicted in Fig. 1.
GRF, CoP, and Force Platforms Moments (FPMs) data were provided at 200 Hz, by using six force platforms embedded in the floor: two force platforms FP4060 (Bertec, Ohio, United States of America), two force platforms FP6090 (Bertec, Ohio, United States of America), and two force platform 9281 EA – FP4060 (Kistler, Winterthur, Switzerland). A threshold of 20 N was used to remove noise from the signals.
The placement of the EMG sensors and retro-reflective markers was always performed by the same assessor, who has several years of experience in this field. The accomplishment of palpation procedures to identify anatomical landmarks was performed according to the standards described by Hermens et al.26 and Tsushima et al.28, to ensure repeatability in the instrumentation procedure, minimizing the occurrence of occasional and systematic errors.
Moreover, all collected data were time-synchronized through a software trigger. More details about the synchronization procedure can be found in the Technical Validation Section.
Experimental protocol
All participants were instructed to perform ten rectilinear walking trials on a 10-meter flat surface with six embedded force platforms (as depicted in Fig. 1), at seven different speeds (1.0, 1.5, 2.0, 2.5, 3.0, 3.5, and 4.0 km/h) controlled with a metronome.
In the beginning, the leg, foot length, and body height were measured with a measuring tape, and the body mass was accessed with a balance. Then, the eight EMG sensors were placed on the intended muscles and their functioning was evaluated. Subsequently, two maximum voluntary contractions (MVCs) were performed for each muscle to normalize the EMG data. With the participant seated and performing 90 degrees between all lower limb joints, two MVCs were done for the VL muscle. The participant performed the maximum knee extension while his/her shank segment was manually immobilized (at 90 degrees with the thigh) by an assessor. For BF muscle, the participants laid on a stretcher, in a ventral decubitus position, and with the knee joint at 30 degrees. With the shank manually immobilized by the same assessor, participants carried out maximum knee flexion. Regarding the TA and GAL muscles, participants laid on a stretcher, in a dorsal decubitus. The manual immobilization of the foot was accomplished while the participant was asked to perform maximum (i) dorsiflexion of the ankle joint (MVC for TA) and (ii) plantar flexion of the ankle joint (MVC for GAL). For all MVCs, the upper body of the participants was immobilized by a second assessor.
Thereafter, the retro-reflective markers were attached to the participant’s body with double-sided adhesive tape. At this point, the participant was ready to start the data acquisition, as illustrated in Fig. 1.
Firstly, the twelve cameras of the motion-capture system were calibrated according to the manufacturer’s instructions, and the force platforms’ functioning was checked. Then, a standing static calibration trial was performed with the arms crossed in front of the chest, looking forward, and with the feet in a comfortable position, aligned with the shoulders. After the calibration procedure, the participant was ready to perform the walking trials. The participants walked comfortably, looking ahead, and taking steps according to the sound of the metronome. The idea was to avoid the participants seeing the force platforms which could lead the participants to inevitably change their walking patterns according to the force platforms’ position. In case the participants reported they had seen the force platforms, the trial was rejected. For coherency during the data collection, only trials in which the participants step on the first force platform with the right foot were admissible. Additionally, every time that the participant performed two contacts in the same force platform, the trial was also rejected. The first imposed walking speed was 1.0 km/h, using a metronome. Each participant walked during a self-selected time to achieve an adequate familiarization level to the speed. After the familiarization period, each participant performed 10 valid walking trials at the controlled gait speed, as presented in Fig. 1. Once recorded 10 trials, the participant rested for five minutes. The walking speed was incremented, and the participant performed a new habituation trial to become acquainted with the new speed (i.e., 1.5 km/h). Ten valid walking trials were registered, the participant rested for five minutes and a new walking speed was imposed (2.0 km/h). This process was repeated for all the speeds.
Dataset elaboration
Once the data acquisition has been performed, an identification method of the Qualisys Track Manager software (Automatic Identification of Markers) was used to preprocess the retro-reflective markers’ coordinates. Despite the retro-reflective markers’ identification method does not allow label duplication, this procedure is not free from defects. Consequently, all marker labeling process was visually inspected by a dedicated assessor. Additionally, to detect marker dropouts, the X, Y, and Z components of each marker trajectory were analyzed. To deal with this phenomenon, the marker trajectory was gap filled through a polynomial interpolation function provided by the Qualisys Track Manager software.
The proposed dataset includes ASCII, *.c3d, and *.mat files containing the raw data, namely EMG signals, 3D marker trajectories, GRFs, FPMs, and coordinates of CoP referred to as the global reference frame of the motion-capture system.
The following procedures were addressed to obtain processed data. First, the joint centers were identified as the midpoint between the medial and lateral retro-reflective markers. For example, the knee joint center corresponds to the medial point between the MK and LK markers. Second, segment masses were determined as fixed proportions of body mass based on built-in anthropometric equations in Visual3D29. Third, we used Visual3D software to determine 3D angles and torques for the lower limb joints and pelvis segment, following the C-Motion modeling recommendations30. Once computed, the 3D angles were low-pass filtered through a 4th order, zero-lag, Butterworth filter set for a cutoff frequency of 6 Hz27. Due to the bidirectional filtering process that minimizes possible phase shifts, data synchronization was not affected. After determining the 3D torques, these data were normalized by the body mass of each participant, being provided the normalized and non-normalized torques. EMG signals were band-pass filtered (4th order, zero-lag, Butterworth filter) with cutoff frequencies of 20 Hz and 450 Hz. Moreover, the envelope of the EMG signals was determined using the Root Mean Square (RMS) value of the signal with a 300 ms movable window, as proposed by Farfán et al.31.
Furthermore, we used a Visual3D software’s function (Automatic Gait Events) for automatic detection of the heel-strike events to split all data into individual gait cycles, where the first sample corresponds to the heel-strike instant. The automatic identification performed by the software was analyzed to find incorrect heel-strike detections. For that, the Z component of the GRF data was analyzed to determine if the heel-strike detection occurred when the Z component started to increase. Every time that an incorrect identification was identified, the heel-strike instant was manually defined. This procedure was useful to gait cycle-normalize to 1001 samples the data of a single gait cycle. During a single trial, the right and left leg performed two and one strides, respectively, since the participants step the first force platform with the right foot. Overall, 1120 trials were collected, containing 1120 and 2240 strides for the left and right leg, respectively. The gait cycle-normalized data, including 3D angles, GRFs, and torques (normalized and non-normalized), filtered, and envelope of the EMG signals for each trial of each participant for the seven walking speeds, are referred to as “processed data” in this dataset, being provided in ASCII and *.mat formats.
Data Records
All data files are available in figshare32. The published data are divided into three folders, namely ASCII files, c3d files, and MAT files folders, representing the three formats available. In each folder, the data are organized participant-by-participant according to the ParticipantID folder naming convention, where ParticipantID represents a folder for each of the sixteen participants that participated in the data collection.
The ASCII files folder contains two other folders (Raw_Data and Processed_Data, with the raw and the processed data saved in the *.txt format). The MAT files folder contains two structs (Raw_Data and Processed_Data with the raw and the processed data saved in the *.mat format and following the same organization as the ASCII files folder). The c3d files folder contains a folder (Raw_Data, with the raw data saved in the *.c3d format and following the same organization as the ASCII files folder). Additionally, these three types of folders also contain a text file (Metadata.txt) containing the metadata of each participant.
It is noteworthy that all data referred to as “raw data” provide information along the 10 m-flat surface. In contrast, the data presented as “processed data” only correspond to the strides carried out in the force platforms, presenting one and two strides for the left and right legs per trial, respectively.
Raw data
Raw data enables future processing procedures, such as filtering or resampling. As this study aims to present the collected data per walking speed for each participant, the Raw_Data folder of the ASCII files and c3d files folders is organized onto seven other folders with the following naming convention: VX, where X can take the values 1, 15, 2, 25, 3, 35, and 4, representing the speeds of 1.0, 1.5, 2.0, 2.5, 3.0, 3.5, and 4.0 km/h, respectively. For each speed, the following six folders are provided: CoP, EMG, GRF, FPM, Trajectories, and Properties, which include the CoP in the force platforms (in m), the EMG signals of the eight muscles (in V), the GRF (in N) and the Moments (in N.m) provided by the force platforms, the trajectories of the 24 retro-reflective markers (in mm), and the units of each data type, respectively. Each one of the first five folders contains the participant’s trials without resampling steps in both ASCII and *.c3d formats. The same organization procedure was followed for the Raw_Data struct of the MAT files folder, presenting the data in the *.mat format.
For both ASCII and *.mat format files, each trial of the CoPs and FPM data present nineteen columns, where the first column represents the trial duration and the remaining eighteen columns correspond to the 3D data from the six force platforms. The first column is labeled as Time and the next eighteen columns are labeled as FPn_P, where (i) n can take the numbers from 1 to 6, corresponding to each one of the six force platforms; and (ii) P represents the 3D plane and it can take the label X, Y, or Z, representing to the sagittal, coronal, and transversal plane, respectively.
For both ASCII and *.mat format files, each EMG data’ trial presents nine columns. The first column represents the trial duration and the remaining columns correspond to the eight monitored muscles of both legs. Thus, the first column is labeled as Time and the subsequent columns are labeled as L_Muscle, where (i) L corresponds to the leg and it can take the nomenclature of R (right leg) or L (left leg); and (ii) Muscle represents the BF, GAL, TA or VL muscles.
For both ASCII and *.mat format files, GRF data’ trials contain seven columns, where the first one represents the trial duration, being labeled as Time, and the columns are labeled as L_GRF_P, where (i) L corresponds to the leg; and (ii) P is regarding the 3D plane. Since we defined a threshold of 20 N, an empty cell in the CoP, FPM, and GRF raw data is created for each timestep, while the threshold is not reached. However, the software tool (Matlab®) that was used to organize and save all data, automatically set the empty cells as NaN values.
At last, for both ASCII and *.mat format files, each trial of the Trajectories data presents the 3D coordinates of each one of the retro-reflective markers presented in Fig. 1(b), labeled as LMarkerP, where L corresponds to the leg, Marker represents each retro-reflective marker and P concerns to the 3D plane. Figure 2(a) illustrates the raw data organization for both ASCII and *.mat formats.
Processed data
The Processed_Data folder of the ASCII files folder, includes the gait cycle-normalized angles and torques (normalized and non-normalized by the participant’s body mass), GRFs, EMG filtered, and EMG envelope data. Its organization method is similar to the Raw_Data folder. The folder of each participant is divided into seven other folders, representing the seven speeds. Each walking speed is split into two new folders, representing the right and left leg. For each leg, the angles (in degrees), torques (N.m), torques normalized by body mass (in N.m/kg), GRF (in N), EMG filtered (in V), and EMG envelope (in % MVC) data are provided per trial in ASCII format files. The units of each data type are presented in the Properties folder. Each trial has the information for the gait-cycle normalization, resampled to 1001 samples. For the identification of strides of the right leg (there are two strides per trial), we used St1 and St2 to represent the data from the first and second strides, respectively. The information of the stride time is presented in the StrideTime folder, in seconds. The same organization procedure was followed for the Processed_Data struct of the MAT files folder, presenting the data in the *.mat format.
For both ASCII and *.mat format files, each trial’s data of the lower limb joint and pelvis angles and normalized and non-normalized torques is organized in a table. The lines represent the samples and each column is labeled considering the following convention: L_Local_P, where (i) L corresponds to the leg; (ii) Local represents each lower limb joint along with the pelvis segment and it can take the name of Ankle, Knee, Hip or Pelvis. The pelvis angles and torques were determined concerning the virtual lab and considering the distal propagation to proximal of the ground reaction forces; and (iii) P is regarding the 3D plane.
For each trial of the GRF data stored in ASCII and *.mat format files, the columns are labeled as L_GRF_P, where (i) L corresponds to the leg; and (ii) P is regarding the 3D plane.
For both filtered EMG and envelope stored in ASCII and *.mat format files, each trial presents eight columns corresponding to the eight monitored muscles of both legs. Each column is labeled as L_Muscle, where (i) L corresponds to the leg; and (ii) Muscle represents the BF, GAL, TA, or VL muscles.
For both ASCII and *.mat format files, the stride time is provided in a text file and a table, respectively. For both, the lines represent the number of trials performed. The first column (Trial) represents the trial number and the second column (St1_Time) represents the stride time in seconds, for the left leg. When considering the right leg, a third column is presented, being labeled as St2_Time. Figure 2(b) represents the Processed Data organization for both ASCII and *.mat formats.
Metadata
The Metadata.txt file contains the following information: (i) ID, the participant’s identification defined as ParticipantX, where X varies from 1 to 16; (ii) Age, the participant’s age in years; (iii) Gender, the participant’s gender (Male or Female, representing male or female, respectively); (iv) Body Height, the participant’s body height in meters; (v) Body Mass, the participant’s body mass in kilograms; (vi) Leg Length, the participant’s leg length in meters; and (vii) Foot Length, the participant’s foot length in meters.
Data limitations
During the dataset organization, we observed some data irregularities in particular gait trials, which might affect the dataset interpretation and use. These trials were removed. Table 2 indicates the number of kept trials considering the participant ID and walking speed for both raw and processed. This information may be considered before the use of the provided data.
Additionally, we identified some general limitations in the dataset. First, the number of participants involved in this study is insufficient to consider the proposed dataset as a reference data for the world young population. Second, the small age range presented by the participants restricts the use of the proposed dataset for a widespread population, covering other age groups. Further, our dataset does not provide EMG data of any hip musculature. Third, this dataset does not include gait transitions, neither other gait activities, such as climbing stairs or ramps since we focused on stable level-ground and continued walking. Thus, the use of the provided gait trajectories as reference trajectories in robotic assistive devices only addresses repetitive level-ground walking therapy at controlled speeds. At last, the data acquisition was conditioned by two external factors: (i) the use of a metronome to impose a cadence, which may change the gait pattern, according to Bertram et al.33; (ii) the requirement of a single step in each force platform.
Technical Validation
Data synchronization
For data synchronization, each data acquisition system was connected via cable connectors to a central computer, running the Qualisys software. When a new recording started, the Qualisys software sent a digital pulse to the EMG and force platform systems, beginning the acquisition in all systems at the same time. The trial recording stopped when another digital pulse was sent to the devices, ending the session at the same instant.
Kinematic, kinetic, and EMG data
Before starting a trial, we addressed the following procedures. First, all cameras were adjusted and calibrated following the manufacturer’s instructions. Second, the force platforms’ functioning was checked, and a reset was performed to remove the offset introduced by the acquisition software.
The retro-reflective markers and the EMG sensors were placed on each participant, as described by Hermens et al.26 and by Rabuffetti et al.27. Before each acquisition session, the quality of the EMG signals was verified, and the sensor’s locations were adjusted when required. If the signal was noisy, the skin was cleaned once again with alcohol. Figure 3 illustrates the averaged muscular activation of the TA, GAL, BF, and VL muscles per speed along with the muscular activations of all trials. Additionally, Fig. 4 depicts the mean of all trials per speed of the sixteen participants for the ankle, knee, and hip joint angles and torques in the three planes. In our study, positive values of angles represent (i) hip and knee flexion, and ankle dorsiflexion in the sagittal plane; (ii) hip and knee adduction, and ankle inversion in the coronal plane; and, (iii) hip and knee internal rotation, and ankle adduction in the transversal plane; while negative values represent (i) hip and knee extension, and ankle plantarflexion in the sagittal plane; (ii) hip and knee abduction, and ankle eversion in the coronal plane; and, (iii) hip and knee external rotation, and ankle abduction in the transversal plane. On the other hand, positive values of torque indicate (i) hip and knee flexion, and ankle dorsiflexion in the sagittal plane; (ii) hip and knee adduction, and ankle inversion in the coronal plane; and, (iii) hip external rotation, knee internal rotation, and ankle adduction in the transversal plane; while negative values represent (i) hip and knee extension, and ankle plantarflexion in the sagittal plane; (ii) hip and knee abduction, and ankle eversion in the coronal plane; and, (iii) hip internal rotation, knee external rotation, and ankle abduction in the transversal plane.
Mean of the ankle, knee, and hip joint angles and torques in the sagittal, coronal, and transversal planes for all participants and trials during a single gait cycle considering the walking speed variation, where: PF – Plantar Flexion; DF – Dorsiflexion; ABD – Abduction; ADD – Adduction; IR – Internal Rotation; ER – External Rotation; EXT – Extension; FLX – Flexion; EVE – Eversion; INV - Inversion.
Comparison with public walking datasets
In order to assess the validity of the proposed dataset, we performed a comparative analysis with a public walking dataset with gait speed conditions as similar as possible to the ones of our dataset. The proposed dataset was compared to Bovi et al.12, which presents a multimodal dataset for a healthy adult population with an age range between 22 and 72 years (43.1 ± 15.4 years). It includes the range of the age of the work herein presented (23.8 ± 2.02 years). Moreover, in Bovi et al.12, the multimodal data was collected for uncontrolled speeds, being categorized as “walking very slowly”, “walking slow”, “walking medium”, “walking fast”, and “natural walking”. The comparative analysis is limited by the differences in the mean value of the age of both populations and the uncertainty of a specific gait speed value. Consequently, we directly compared data for walking speeds of 4.0 km/h with data acquired for a “natural walking” of the public dataset (Bovi et al.12) since 4.0 km/h commonly represents a human natural gait speed22. The determined Pearson’s correlation ranged from 0.675 to 0.986 and from 0.696 to 0.993 for joint angles and torques, respectively. EMG signals from BF and TA muscles (the muscles in common) presented Pearson’s correlation of 0.828 and 0.724, respectively.
Furthermore, we compared the proposed dataset with the one presented by D. Winter20, regarding the effects of walking speed variation in walking dynamics. In D. Winter20, the lower limb joint torques were collected from an adult population with a mean age of 25.6 ± 6.2 years, a mean body mass of 69.1 ± 8.8 kg, and a mean body height of 1.75 ± 0.078 m. The collected data were analyzed for three self-selected walking speeds (slow, natural, and fast), in the sagittal plane. In our study, we verified that as the walking speed increases, the ankle dorsiflexion torque, the knee flexion torque, and the hip extension torque also increased. These findings were also reported by D. Winter20, conferring robustness to the proposed multimodal dataset. Further, our study was also compared to Fukuchi et al.19, in which 3D lower limb joint kinematics and kinetics were collected from 42 adult volunteers: 24 young adults with a mean age of 27.6 ± 4.4 years, a mean body height of 171.1 ± 10.5 cm, and mean body mass of 68.4 ± 12.2 kg, and 18 older adults with a mean age of 62.7 ± 8.0 years, a mean body height of 1.62 ± 0.095 m, and a mean body mass of 66.9 ± 10.1 kg. Data were collected at three self-selected walking speeds (slow, natural, and fast) during overground walking. It was found a coherency between the proposed dataset and the one presented by Fukuchi et al.19 concerning the effects caused by the walking speed in the gait dynamics. Nonetheless, in our study, the behavior of the sagittal and transversal ankle joint angles, and the coronal and transversal knee joint angles in response to the walking speed variation was different from the one reported by Lelas et al.13, Fukuchi et al.19, and D. Winter20. These findings may be justified by the highly reduced walking speeds innovatively approached in our study. Data collection of all covered speeds followed standards of human gait data acquisition, ensuring the reliability of the observed lower limb joint patterns.
Code availability
A Matlab® script (CodeAvailability.m available in figshare32) is provided to demonstrate how the dataset can be accessed and how to visualize the processed knee joint angles from a single participant (in this case, Participant 1) in the sagittal plane, for the walking speed of 4 km/h. Moreover, the script also presents an example of creating the graph of the vertical component of the raw GRF for a walking speed of 3 km/h. Additionally, it is provided a simple segment of code to access the stride time of all trials for a speed of 2.5 km/h. To run the provided code, the script must be placed on a folder that contains the MAT files folder.
References
Perry, J. Gait Analysis: Normal and Pathological Function. (SLACK Incorporated, 1992).
Jang, S. H. The recovery of walking in stroke patients: a review. Int. J. Rehabil. Res. 33, 285–289 (2010).
Campos Figueiredo, J. S. Smart Wearable Orthosis to Assist Impaired Human Walking. (University of Minho, 2019).
Koopman, B., van Asseldonk, E. H. F. & van der Kooij, H. Speed-dependent reference joint trajectory generation for robotic gait support. J. Biomech. 47, 1447–1458 (2014).
Lencioni, T., Carpinella, I., Rabuffetti, M., Marzegan, A. & Ferrarin, M. Human kinematic, kinetic and EMG data during different walking and stair ascending and descending tasks. Sci. Data 6 (2019).
Winter, D. A. Biomechanics and Motor Control of Human Movement. Biomechanics and Motor Control of Human Movement: Fourth Edition (John Wiley & Sons, Inc., 2009).
Rao, A. K., Muratori, L., Louis, E. D., Moskowitz, C. B. & Marder, K. S. Spectrum of gait impairments in presymptomatic and symptomatic Huntington’s disease. Mov. Disord. 23, 1100–1107 (2008).
Gage, J. R., Schwartz, M. H., Koop, S. E. & Novacheck, T. F. The Identification And Treatment Of Gait Problems In Cerebral Palsy. Archives of Pediatrics & Adolescent Medicine (London: Mac Keith Press, 2009).
Carpinella, I. et al. Locomotor function in the early stage of Parkinson’s disease. IEEE Trans. Neural Syst. Rehabil. Eng. 15, 543–551 (2007).
Beaman, C. B., Peterson, C. L., Neptune, R. R. & Kautz, S. A. Differences in self-selected and fastest-comfortable walking in post-stroke hemiparetic persons. Gait Posture 31, 311–316 (2010).
Stoquart, G., Detrembleur, C. & Lejeune, T. Effect of speed on kinematic, kinetic, electromyographic and energetic reference values during treadmill walking. Neurophysiol. Clin. Neurophysiol. 38, 105–116 (2008).
Bovi, G., Rabuffetti, M., Mazzoleni, P. & Ferrarin, M. A multiple-task gait analysis approach: kinematic, kinetic and EMG reference data for healthy young and adult subjects. Gait Posture 33, 6–13 (2011).
Lelas, J. L., Merriman, G. J., Riley, P. O. & Kerrigan, D. C. Predicting peak kinematic and kinetic parameters from gait speed. J. Gait Posture 17, 106–112 (2003).
Schwartz, M. H., Rozumalski, A. & Trost, J. P. The effect of walking speed on the gait of typically developing children. J. Biomech. 41, 1639–1650 (2008).
Branco, M., Santos-Rocha, R. & Vieira, F. Biomechanics of gait during Pregnancy. Sci. World J. 1–5 (2014).
DeVita, P. & Hortobágyi, T. Obesity is not associated with increased knee joint torque and power during level walking. J. Biomech. 36, 1355–1362 (2003).
Quesada, P. M., Mengelkoch, L. J., Hale, R. C. & Simon, S. R. Biomechanical and metabolic effects of varying backpack loading on simulated marching. Ergonomics 43, 293–309 (2000).
Van Den Bogert, A. J., Geijtenbeek, T., Even-Zohar, O., Steenbrink, F. & Hardin, E. C. A real-time system for biomechanical analysis of human movement and muscle function. Med. Biol. Eng. Comput. 51, 1069–1077 (2013).
Fukuchi, C. A., Fukuchi, R. K. & Duarte, M. A public dataset of overground and treadmill walking kinematics and kinetics in healthy individuals. PeerJ 1–17 (2018).
Winter, D. A. Kinematic and kinetic patterns in human gait: Variability and compensating effects. Hum. Mov. Sci. 3, 51–76 (1984).
Moore, J. K., Hnat, S. K. & van den Bogert, A. J. An elaborate data set on human gait and the effect of mechanical perturbations. PeerJ. (2015).
Hanlon, M. & Anderson, R. Prediction methods to account for the effect of gait speed on lower limb angular kinematics. Gait Posture 24, 280–287 (2006).
Cho, S. H., Park, J. M. & Kwon, O. Y. Gender differences in three dimensional gait analysis data from 98 healthy Korean adults. Clin. Biomech. 19, 145–152 (2004).
Kerrigan, D. C., Todd, M. & Croce, U. Gender differences in joint biomechanics during walking. Am. J. Phys. Med. Rehabil. 77, 2–7 (1998).
Perkins, J. M., Subramanian, S. V., Davey Smith, G. & Özaltin, E. Adult height, nutrition, and population health. Nutr. Rev. 74, 149–165 (2016).
Hermens, H. J., Freriks, B., Disselhorst-Klug, C. & Rau, G. Development of recommendations for SEMG sensors and sensor placement procedures. J. Electromyogr. Kinesiol. 10, 361–374 (2000).
Rabuffetti, M. et al. The LAMB gait analysis protocol: Definition and experimental assessment of operator-related variability. Proc. Inst. Mech. Eng. Part H J. Eng. Med. 233, 342–353 (2019).
Tsushima, H., Morris, M. E. & McGinley, J. Test-Retest Reliability and Inter-Tester Reliability of Kinematic Data from a Three-Dimensional Gait Analysis System. J. Japanese Phys. Ther. Assoc. 6, 9–17 (2003).
C-Motion. Segment Mass. MediaWiki https://www.c-motion.com/v3dwiki/index.php?title=Segment_Mass (2016).
C-Motion. Visual3D Overview. MediaWiki https://www.c-motion.com/v3dwiki/index.php?title=Visual3D_Overview (2019).
Farfán, F. D., Politti, J. C. & Felice, C. J. Evaluation of EMG processing techniques using Information Theory. Biomed. Eng. Online 9 (2010).
Moreira, L., Figueiredo, J., Fonseca, P., Vilas-Boas, J. P. & Santos, C. P. Lower limb kinematic, kinetic, and EMG data from young healthy humans during walking at controlled speeds. figshare https://doi.org/10.6084/m9.figshare.c.4923162 (2020).
Bertram, J. E. A. & Ruina, A. Multiple Walking Speed–frequency Relations are Predicted by Constrained Optimization. J. Theor. Biol. 209, 445–453 (2001).
Acknowledgements
We thank Alexandros Stamoulis and Tjasa Zorn for the assistance with the experiments. This work has been supported in part by the Fundação para a Ciência e Tecnologia (FCT) with the Reference Scholarship under Grant 2020.05711.BD, and in part by the FEDER Funds through the COMPETE 2020—Programa Operacional Competitividade e Internacionalização (POCI) and P2020 with the Reference Project SmartOs Grant POCI-01-0247-FEDER-039868, and by FCT national funds, under the national support to R&D units grant, through the reference project UIDB/04436/2020 and UIDP/04436/2020.
Author information
Authors and Affiliations
Contributions
L.M. wrote the manuscript, conceived data collection, and contributed to the elaboration of the software algorithm to organize the proposed database. J.F. contributed to data collection, analysis, and edition of the manuscript. P.F. contributed to data collection and software development to process the collected data. J.P.V. supervised the study. C.P.S. conceived and supervised the study, and contributed to the edition of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
The Creative Commons Public Domain Dedication waiver http://creativecommons.org/publicdomain/zero/1.0/ applies to the metadata files associated with this article.
About this article
Cite this article
Moreira, L., Figueiredo, J., Fonseca, P. et al. Lower limb kinematic, kinetic, and EMG data from young healthy humans during walking at controlled speeds. Sci Data 8, 103 (2021). https://doi.org/10.1038/s41597-021-00881-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-021-00881-3
This article is cited by
-
Robust and novel attention guided MultiResUnet model for 3D ground reaction force and moment prediction from foot kinematics
Neural Computing and Applications (2024)
-
Surface electromyogram, kinematic, and kinetic dataset of lower limb walking for movement intent recognition
Scientific Data (2023)
-
A kinematic, imaging and electromyography dataset for human muscular manipulability index prediction
Scientific Data (2023)
-
NONAN GaitPrint: An IMU gait database of healthy young adults
Scientific Data (2023)
-
High-density EMG, IMU, kinetic, and kinematic open-source data for comprehensive locomotion activities
Scientific Data (2023)