Abstract
Species distribution records are a prerequisite to follow climate-induced range shifts across space and time. However, synthesizing information from various sources such as peer-reviewed literature, herbaria, digital repositories and citizen science initiatives is not only costly and time consuming, but also challenging, as data may contain thematic and taxonomic errors and generally lack standardized formats. We address this gap for important marine ecosystem-structuring species of large brown algae and seagrasses. We gathered distribution records from various sources and provide a fine-tuned dataset with ~2.8 million dereplicated records, taxonomically standardized for 682 species, and considering important physiological and biogeographical traits. Specifically, a flagging system was implemented to signal potentially incorrect records reported on land, in regions with limiting light conditions for photosynthesis, and outside the known distribution of species, as inferred from the most recent published literature. We document the procedure and provide a dataset in tabular format based on Darwin Core Standard (DwC), alongside with a set of functions in R language for data management and visualization.
Measurement(s) | Species • Distribution |
Technology Type(s) | digital curation |
Factor Type(s) | geographic location |
Sample Characteristic - Organism | Fucales • Laminariales • Tilopteridales • Cymodoceaceae • Hydrocharitaceae • Posidoniaceae • Zosteraceae |
Sample Characteristic - Environment | marine biome |
Sample Characteristic - Location | global |
Machine-accessible metadata file describing the reported data: https://doi.org/10.6084/m9.figshare.12011523
Similar content being viewed by others
Background & Summary
Bioclimatic modelling1,2, macroecology3 and evolution4 are fields that have recently seen a boost in broad scale analyses owing to increased accessibility of large scale biodiversity data. Although these can be obtained from digital online databases (e.g., GBIF, the Global Biodiversity Information Facility, www.gbif.org and OBIS, the Ocean Biogeographic Information System, www.obis.org), herbarium (e.g., Macroalgal Herbarium Portal, www.macroalgae.org), museum collections, as well as citizen science initiatives5,6,7, they can be very incomplete and contain geographical and taxonomic errors. In particular, studies focused on the impacts of global climate changes8,9, or locating evolutionary biodiversity hotspots10,11, require complete and extremely accurate baselines on the distribution of species across space and time12.
Collating broad-scale biodiversity data from multiple sources is challenged by two major obstacles. First, the lack of complete database compatibility allowing efficient information exchange between distinct sources, alongside with inconsistent file structures13,14, leaves data frequently scattered, even for well‐known taxa15. Second, the quality of several sources has been questioned regarding potential geographical data errors16. This is a serious limitation since unreliable biased records can deeply influence the outcomes of research analyses. For instance, distribution models can be strongly influenced by particular marginal records. While records of marine species falling on land (and vice-versa) can be easily identified and dealt with10, those distributed in climatically unfavorable regions (i.e., outside species’ niche), beyond range margins or dispersal capacities, should be verified and corrected when necessary. Wrong records may be even more likely for rare, elusive, or cryptic species that can be easily confused with others, more common and broadly distributed17. An additional problem that is more evident and easier to tackle is related to taxonomic data errors16, which can deeply confound the baseline of a species’ distribution18. When properly reviewed, databases can integrate quality control flags to identify potential data limitations. While some research communities have developed quality control standards on data (e.g., The Ocean Data Standards and Best Practices Project, www.oceandatastandards.org), no implementation has been done so far for the aforementioned data limitations, even in major online data sources providing large scale biodiversity data.
Here we provide a fine-tuned dataset of marine forests at global scales, with occurrence records gathered from numerous independent sources19,20 and flagged with automatic and manual pipelines to increase data reliability in terms of geographical (including depth) and taxonomical traits. “Marine forests” is a common name used here to designate large brown algae (kelp and fucoids) and seagrasses. These blue-green infrastructures rank among the most productive and biodiversity-rich ecosystems21, supporting diverse food webs22,23, critical habitats and nursery grounds for numerous associated species24,25. They increase local biodiversity levels23,25,26,27 and provide key ecological services21 such as nutrient cycling, carbon sequestration28,29, sediment stabilization, and natural protection against ocean wave energy23. Because climate change is shifting their distribution and abundance worldwide1,8,30,31, a comprehensive dataset providing essential baselines is needed to better report and understand marine forests’ variability across space and time14.
Methods
Data compilation
Occurrence records of marine forests of large brown algae (orders Fucales, Laminariales and Tilopteridales), and seagrasses (families Cymodoceaceae, Hydrocharitaceae, Posidoniaceae and Zosteraceae) were gathered from online repositories and herbaria, peer-reviewed scientific literature and citizen science initiatives with independently verifiable data (e.g., supported by photos). Only records with no copyright for any use and without any restriction (e.g., CC0, www.creativecommons.org), or any use with appropriate attribution (e.g., CC BY), were stored in the dataset (please refer to the analytical list of data sources; Suppl. Table 1).
Data treatment
The dataset structure was based on Darwin Core Standard (DwC)32. This framework for biodiversity data offers a stable and flexible framework to store all fields available in original data sources. Moreover, it provides standard identifiers, labels, and definitions, allowing a full link-back to original data sources.
Taxonomic standardization was performed with the World Register of Marine Species (WoRMS; www.marinespecies.org), a universally authoritative open-access reference system for marine organisms. This tool provides a unique identifier (aphiaID) that enabled to link each taxon originally captured, to an internationally accepted standardized name with associated taxonomic information (including hierarchy, rank, acceptance status and synonymy) that will continue to be updated in the future in case of taxonomic or name changes. In the rare cases of no match with WoRMS (including misspelled entries), or uncertain taxonomic status, the records were removed from the dataset.
Geographical locations were available for most records as coordinates in decimal degrees. For those records missing coordinates, but including information on location, an automatic geocoding procedure was performed with OpenStreetMap33,34 service (http://planet.openstreetmap.org).
Since unique records may be available across distinct data sources, the final aggregated dataset was subjected to the removal of duplicate records. These were considered when belonging to the same taxon, and recorded in the same exact geographical location (longitude, latitude and depth) and date (year, month and day).
Quality control
To achieve a fine-tuned dataset, a flagging system was implemented to identify records with doubtful geographical and depth locations. This started by flagging records occurring on land, by using a 1 km threshold from shoreline. This distance represented the lower spatial resolution of the polygon used to define landmass (OpenStreetMap geographic information33). Light availability for photosynthesis was further considered, since it is the main environmental driver restricting the vertical distribution of marine forests35. Limiting light was favored in detriment of bathymetry, because it varies with depth throughout the global ocean, particularly in oceanic regions, were it reaches deeper waters1. Available light at bottom was extracted from Bio-ORACLE36, a dataset providing benthic environmental layers (i.e., along the bottom of the ocean). Because Bio-ORACLE layers are available for 3 different depth ranges, the maximum light value per record was chosen as a conservative approach to estimate the potential depth range for a given location. Records were flagged when light values were below the known limiting threshold of 50 E.m−2.year−1 for marine forests’ photosynthesis35,37. This flag was not applied to the brown algae Sargassum fluitans, Sargassum natans38 and Sargassum pusillum39 as they can complete a full life cycle floating on the sea surface.
Finally, all records were manually verified to identify potential outliers outside the known distribution of species. This information was based on the most recent published literature and by consulting experts when possible. Because distributional ranges are often documented at an administrative level (e.g., country), the flagging procedure integrated the Marine Ecoregions of the World (MEOW)40, a scheme that represents the broad-scale distributional patterns of species/communities in the ocean40. Records were flagged when distributed in a MEOW region not considered in the information available in the literature or provided by experts. The MEOW has 3 distinct levels dividing the globe into 12 realms, 62 provinces and 232 ecoregions40. We adopted the intermediate level “provinces” to reduce commission errors (cases incorrectly identified as potential outliers) and omission errors (outliers left out, or omitted), potentially arising while considering “realms” and “ecoregions”, respectively. Records were removed from the database when no information was available in literature to support the actual distribution of species.
Data Records
The dataset is publicly accessible for download in a permanent Figshare41 repository (https://doi.org/10.6084/m9.figshare.7854767). A version containing only pruned records is also accessible at https://www.dataone.org and https://www.marineforests.com.
Taxonomic coverage
The dataset provided41 covers 682 accepted taxa (at the species level; Suppl. Table 2) belonging to the orders Fucales, Laminariales and Tilopteridales (i.e., brown macroalgae; Fig. 1), and the families Cymodoceaceae, Hydrocharitaceae, Posidoniaceae and Zosteraceae (i.e., seagrass; Fig. 2).
Spatial and temporal coverage
The dataset contains 2,751,805 records of occurrence (brown algae: 1,088,448; seagrasses: 1,663,357; Table 1) globally distributed between the years 1663 and 2018 (Fig. 3), increasing by 47.43% the data available in the two major online repositories GBIF and OBIS (Figs. 4 and 5).
Data collection sources
The dataset gathered information from 18 distinct repositories, 15 herbaria and 569 literature sources. The majority of records resulted from external repositories (82.56% of records), followed by literature (16.07% of records) and herbaria (1.35% of records; Table 1). The main repositories GBIF and OBIS accounted for 52.57% of all records). In terms of species number, the main sources of data were external repositories, followed by herbaria and literature. These covered 96.77%, 61.14% and 13.04% of species, respectively (Table 2).
Technical Validation
The dataset gathered information from multiple sources, some of which may be automatically interoperable, sharing erratic duplicated data, regardless of the credibility of the source. These data can be used in scientific studies, potentially generating misleading results. To address the challenge, we developed a specific quality control data treatment based on automatic and manual pipelines.
The taxonomic standardization using WORMS discarded any misspelled or no-match entries from the dataset, and aggregated 1116 initial taxa into 682 accepted taxa (at the species level). As new taxa are being described and their current status is constantly changing, WoRMS may not yet contain all updated statuses42, however, it is continuously being improved and is considered the best available source for marine taxonomic standardization. Together with the identification of duplicate entries, records missing coordinate information or information regarding species’ distributional ranges, our approach removed 2,676,350 initial entries from the dataset.
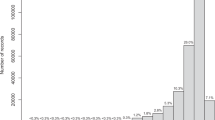
The automatic flagging procedure identified 1.21% of records located on land, and an additional 6.88% records without suitable light conditions for photosynthesis (Table 1). The manual verification based on published literature and consulting experts flagged 2.74% of records as potential outliers outside the know distribution of species (75,369 records; Table 1). Considering the three flags implemented, literature records appeared the least biased (unique exception of literature records for seagrasses flagged over land; Table 1), followed by digital repositories and herbaria (Table 2). The number of species flagged by manual verification against known distributional ranges was the lowest for literature (26.96%), followed by repositories (36.96%) and herbaria (60.43%; Table 2).
The flagging system implemented, not available in any of the 33 repositories and herbaria consulted, allowed delivering a fine-tuned dataset of 2,485,534 georeferenced records gathered from multiple sources, with no taxonomic errors (based on the WoRMS current information), no duplicate entries, no records in unsuitable habitats (i.e. land or low light conditions) or too distant from species’ biogeographical ranges.
The use of a flagging system allowed retaining valuable data that should not be discarded. For instance, some large brown algae and seagrasses can often be found as rafts43, floating on the sea surface, hundreds of kilometers away from their original source44,45. While these records are not particularly suitable to build ecological models aimed for benthic species, they are highly valuable to address dispersal ecology. Instead of considering such cases as outliers for exclusion, flagging allows keeping records for users to decide their final use.
The dataset will continue to receive new data records from its multiple sources, as new literature gets published and new observations are made. Taxonomic and error corrections will continuously be made over the years, from experts (ecologists, taxonomists and naturalists), allowing continuous flagging of doubtful records.
R functions for data management and visualization
In addition to the dataset, we developed a set of functions in R language (R Development Core Team, 2018) to facilitate extraction, listing and visualization of occurrence records (e.g., function to export data as geospatial vectors for geographic information systems). All functions are detailed in Table 3 and can be easily installed by entering the following line into the command prompt:
source(“https://raw.githubusercontent.com/jorgeassis/marineforestsDB/master/sourceMe.R”).
Usage Notes
The dataset follows the FAIR principle of Findability, Accessibility, Interoperability and Reusability of data46. It is made available as two distinct files in tabular format. The first aggregates all data with no taxonomic errors and no duplicate entries and includes the three fields implemented to flag records. The additional file provides a pruned version of the dataset discarding all potentially biased records.
The dataset complies with Darwin Core Standard (DwC)32, providing information on taxonomy, geographical location (e.g., coordinates in decimal degrees, depth and uncertainty), reference to original sources (including permanent identifiers; bibliographic Citation DOI), as well as the flagging system implemented (Table 4).
The integration of the dataset with a set of functions in R language allows easy data acquisition and smooth integration with already available statistical tools, such as those aiming for Ecological Niche Modeling47,48. For instance, the dataset can be used to describe the global distribution of species12,49, address niche-based questions3,50,51, support biodiversity and ecosystem-based conservation10,52,53, and to understand correlations between anthropogenic pressures and population extinctions54. Additionally, the availability of standard data layers delivering past and future climate change scenarios36,55 may further expand the applications of this dataset to predict range shifts9,56,57 or hypothesize important evolutionary scenarios, such as mapping climate-refugia where higher and endemic biodiversity evolved43,58,59.
Data transparency and accuracy is a prerequisite for avoiding flawed and/or misleading conclusions, especially when provided to stakeholders and decision makers. The pipelines implemented are explicit, ensuring the clarity and reproducibility of the process and contributing to public data in standard formats (i.e., the Darwin Core Standard). With the flagging system, users can fine-tune the original dataset according to their research needs and boost the quality of their results. Particularly, when requested by decision-makers, more accurate outcomes may provide important climate change-integrated conservation strategies60, as well as feed important baseline assessments, like those required in the scope of the Intergovernmental Science-Policy Platform on Biodiversity and Ecosystem Services (IPBES).
Code availability
Data management was performed using R computing language61. The functions developed to manage and flag the dataset are permanently available in a Github repository (https://github.com/jorgeassis/marineforestsDB).
References
Assis, J., Araújo, M. B. & Serrão, E. A. Projected climate changes threaten ancient refugia of kelp forests in the North Atlantic. Glob. Chang. Biol. 24, 1365–2486 (2017).
Thuiller, W. Patterns and uncertainties of species’ range shifts under climate change. Glob. Chang. Biol. 10, 2020–2027 (2004).
Verbruggen, H. et al. Macroecology meets macroevolution: Evolutionary niche dynamics in the seaweed Halimeda. Glob. Ecol. Biogeogr. 18, 393–405 (2009).
Waltari, E. & Hickerson, M. J. Late Pleistocene species distribution modelling of North Atlantic intertidal invertebrates. J. Biogeogr. 40, 249–260 (2013).
Azzurro, E., Broglio, E., Maynou, F. & Bariche, M. Citizen science detects the undetected: the case of Abudefduf saxatilis from the Mediterranean Sea. Manag. Biol. Invasions 4, 167–170 (2013).
Cox, J. et al. Defining and Measuring Success in Online Citizen Science: A Case Study of Zooniverse Projects. Comput. Sci. Eng. 17, 28–41 (2015).
Assis, J. et al. Findkelp, a GIS-based community participation project to assess Portuguese kelp conservation status. J. Coast. Res. 3, 1469–1473 (2009).
Assis, J., Lucas, A. V., Bárbara, I. & Serrão, E. Á. Future climate change is predicted to shift long-term persistence zones in the cold-temperate kelp Laminaria hyperborea. Mar. Environ. Res. 113, 174–182 (2016).
Neiva, J. et al. Genes Left Behind: Climate Change Threatens Cryptic Genetic Diversity in the Canopy-Forming Seaweed Bifurcaria bifurcata. PLoS One 10, e0131530 (2015).
Boavida, J., Assis, J., Silva, I. & Serrão, E. A. Overlooked habitat of a vulnerable gorgonian revealed in the Mediterranean and Eastern Atlantic by ecological niche modelling. Sci. Rep. 6, 36460 (2016).
Assis, J. et al. Deep reefs are climatic refugia for genetic diversity of marine forests. J. Biogeogr. 43, 833–844 (2016).
Chefaoui, R. M., Assis, J., Duarte, C. M. & Serrão, E. A. Large-Scale Prediction of Seagrass Distribution Integrating Landscape Metrics and Environmental Factors: The Case of Cymodocea nodosa (Mediterranean–Atlantic). Estuaries and Coasts 39, 123–137 (2015).
Shanmughavel, P. An overview on biodiversity information in databases. Bioinformation 1, 367–369 (2007).
Duputié, A., Zimmermann, N. E. & Chuine, I. Where are the wild things? Why we need better data on species distribution. Glob. Ecol. Biogeogr. 23, 457–467 (2014).
Yesson, C. et al. How global is the global biodiversity information facility? PLoS One 2, e1124 (2007).
Morris, R. A. Encyclopedia of Biodiversity: Second Edition. Academic Press (Princeton University, 2013).
Aubry, K. B., Raley, C. M. & McKelvey, K. S. The importance of data quality for generating reliable distribution models for rare, elusive, and cryptic species. PLoS One 12, e0179152 (2017).
Beck, J., Böller, M., Erhardt, A. & Schwanghart, W. Spatial bias in the GBIF database and its effect on modeling species’ geographic distributions. Ecol. Inform. 19, 1–10 (2014).
Ceccarelli, S. et al. Data Descriptor: DataTri, a database of American triatomine species occurrence. Sci. Data 24, 180071 (2018).
Dyer, E. E., Redding, D. W. & Blackburn, T. M. The global avian invasions atlas, a database of alien bird distributions worldwide. Sci. Data 4, 170041 (2017).
Costanza, R. et al. The value of the world’s ecosystem services and natural capital. Nature 387, 253–260 (1998).
Araújo, R. M. et al. Status, trends and drivers of kelp forests in Europe: an expert assessment. Biodivers. Conserv. 25, 1319–1348 (2016).
Green, E. P. & Short, F. T. World Atlas Seagrass. (University of California Press, Berkeley, USA, 2003).
Hemminga, M. A. & Duarte, C. M. Seagrass Ecology. (Cambridge University Press, 2000).
Christie, H., Norderhaug, K. M. & Fredriksen, S. Macrophytes as habitat for fauna. Mar. Ecol. Prog. Ser. 396, 221–233 (2009).
Borg, J. A., Rowden, A. A., Attrill, M. J., Schembri, P. J. & Jones, M. B. Wanted dead or alive: High diversity of macroinvertebrates associated with living and ‘dead’ Posidonia oceanica matte. Mar. Biol. 149, 667–677 (2006).
Reaka-Kudla, M. L. The Global Biodiversity of Coral Reefs: A Comparison with Rain Forests. In Biodiversity II: Understanding and Protecting Our Biological Resources (eds. Reaka-Kudla, M. L., Wilson, D. E. & Wilson, E. O.) 83–108 (Joseph Henry Press, 1997).
Fourqurean, J. W. et al. Seagrass ecosystems as a globally significant carbon stock. Nat. Geosci. 5, 505–509 (2012).
Chung, I. K. et al. adaptation against global warming: Korean Project Overview. ICES J. Mar. Sci. 68, 66–74 (2012).
Neiva, J. et al. Climate Oscillations, Range Shifts and Phylogeographic Patterns of North Atlantic Fucaceae. In Seaweed Phylogeography (eds. Hu, Z.-M. & Fraser, C.) 279–308 (Springer Netherlands, 2016).
Assis, J. et al. Major shifts at the range edge of marine forests: the combined effects of climate changes and limited dispersal. Sci. Rep. 7(44348), 1–10 (2017).
Wieczorek, J. et al. Darwin core: An evolving community-developed biodiversity data standard. PLoS One 7, e29715 (2012).
Haklay, M. & Weber, P. OpenStreet map: User-generated street maps. IEEE Pervasive Comput. 1, 12–18 (2008).
Contributors, O. Openstreetmap. Retrieved from, https://planet.openstreetmap.org (2015).
Graham, M. H., Kinlan, B. P., Druehl, L. D., Garske, L. E. & Banks, S. Deep-water kelp refugia as potential hotspots of tropical marine diversity and productivity. Proc. Natl. Acad. Sci. USA 104, 16576–16580 (2007).
Assis, J. et al. Bio-ORACLE v2.0: Extending marine data layers for bioclimatic modelling. Glob. Ecol. Biogeogr. 27, 277–284 (2017).
Vaquer-Sunyer, R. & Duarte, C. M. Thresholds of hypoxia for marine biodiversity. Proc. Natl. Acad. Sci. 105, 15452–15457 (2008).
Amaral-Zettler, L. A. et al. Comparative mitochondrial and chloroplast genomics of a genetically distinct form of Sargassum contributing to recent “Golden Tides” in the Western Atlantic. Ecol. Evol. 7, 516–525 (2017).
Taylor, W. R. A pelagic Sargassum from the Western Atlantic. Contr. Univ. Mich, Herb. 27, 73–75 (1975).
Spalding, M. D. et al. Marine Ecoregions of the World: A Bioregionalization of Coastal and Shelf Areas. Bioscience 57, 573–583 (2007).
Assis, J. et al. A fine-tuned global distribution dataset of marine forests. figshare https://doi.org/10.6084/m9.figshare.7854767 (2019).
Costello, M. J. et al. Global Coordination and Standardisation in Marine Biodiversity through the World Register of Marine Species (WoRMS) and Related Databases. 8 (2013).
Waters, J. M., King, T. M., Fraser, C. I. & Craw, D. Crossing the front: Contrasting stormforced dispersal dynamics revealed by biological, geological and genetic analysis of beach-cast kelp. J. R. Soc. Interface 15 (2018).
Assis, J. et al. Past climate changes and strong oceanographic barriers structured low - latitude genetic relics for the golden kelp Laminaria ochroleuca. 45, 2326–2336 (2018).
Thiel, M. & Haye, P. A. The ecology of rafting in the marine environment. iii. Biogeographical and evolutionary consequences. Oceanogr. Mar. Biol. An Annu. Rev. 44, 323–429 (2006).
Wilkinson, M. D. et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 3, 160018 (2016).
Naimi, B. & Araújo, M. B. sdm: a reproducible and extensible R platform for species distribution modelling. Ecography (Cop.). 39, 368–375 (2016).
Thuiller, W., Lafourcade, B., Engler, R. & Araújo, M. B. BIOMOD - A platform for ensemble forecasting of species distributions. Ecography (Cop.). 32, 369–373 (2009).
Chaudhary, C., Saeedi, H. & Costello, M. J. Bimodality of Latitudinal Gradients in Marine Species Richness. Trends Ecol. Evol. 31, 670–676 (2017).
Assis, J. et al. Oceanographic Conditions Limit the Spread of a Marine Invader along Southern African Shores. PLoS One 10, e0128124 (2015).
Lee-Yaw, J. A. et al. A synthesis of transplant experiments and ecological niche models suggests that range limits are often niche limits. Ecol. Lett. 19, 710–722 (2016).
Guisan, A. & Thuiller, W. Predicting species distribution: Offering more than simple habitat models. Ecol. Lett. 8, 993–1009 (2005).
Guisan, A. et al. Predicting species distributions for conservation decisions. Ecol. Lett. 16, 1424–1435 (2013).
Scherner, F. et al. Coastal urbanization leads to remarkable seaweed species loss and community shifts along the SW Atlantic. Mar. Pollut. Bull. 76, 106–115 (2013).
Moss, R. H. et al. The next generation of scenarios for climate change research and assessment. Nature 463, 747–756 (2010).
Burrows, M. T. et al. Geographical limits to species-range shifts are suggested by climate velocity. Nature 507, 492–5 (2014).
Martínez, B. et al. Distribution models predict large contractions of habitat-forming seaweeds in response to ocean warming. Divers. Distrib. 24, 1350–1366 (2018).
Waltari, E. et al. Locating pleistocene refugia: Comparing phylogeographic and ecological niche model predictions. PLoS One 2, e563 (2007).
Assis, J., Serrão, E. A., Claro, B., Perrin, C. & Pearson, G. A. Climate-driven range shifts explain the distribution of extant gene pools and predict future loss of unique lineages in a marine brown alga. Mol. Ecol. 23, 2797–2810 (2014).
Hannah, L., Midgley, G. F. & Millar, D. Climate change-integrated conservation strategies. Glob. Ecol. Biogeogr. 11, 485–495 (2002).
R Development Core Team. R: A Language and Environment for Statistical Computing. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2016).
GBIF.org, Global Biodiversity Information Facility Home Page, https://www.gbif.org (2019).
OBIS: Ocean Biogeographic Information System Home Page, https://www.obis.org (2019).
Core, D. Darwin Core maintenance group, Biodiversity Information Standards (TDWG). Zenodo 1 (2014).
Acknowledgements
This study was supported by a Pew Marine Fellowship (EAS), the European Maritime and Fisheries Fund and the MAR2020 program through project REDAMP (MAR-01.04.02-FEAMP-0015), and the Foundation for Science and Technology (FCT) of Portugal through fellowships to J.A. (SFRH/BPD/111003/2015), D.A. (SFRH/BPD/95334/2013), EAS (SFRH/BSAB/150485/2019) and E.F. (SFRH/BD/144878/2019), the transitional norm - DL57/2016/CP1361/CT0035 and D.L. 57/2016/CP1361/CT0036, and projects GENEKELP (PTDC/MAR-EST/6053/2014), MARFOR (BIODIVERSA/004/2015) and UIDB/04326/2020. We thank all the many taxonomy experts and citizen science volunteers that verified and contributed verifiable data records.
Author information
Authors and Affiliations
Contributions
J.A. and E.A.S. conceived the study. J.A., E.F. and A.O. designed the data pipelines. D.F., J.N., S.F. and E.A.S. revised the data and queried taxonomic experts about doubtful records. D.A. contributed funds and tools. All authors wrote and reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
The Creative Commons Public Domain Dedication waiver http://creativecommons.org/publicdomain/zero/1.0/ applies to the metadata files associated with this article.
About this article
Cite this article
Assis, J., Fragkopoulou, E., Frade, D. et al. A fine-tuned global distribution dataset of marine forests. Sci Data 7, 119 (2020). https://doi.org/10.1038/s41597-020-0459-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-020-0459-x
This article is cited by
-
A review of the current potential of European brown seaweed for the production of biofuels
Energy, Sustainability and Society (2024)
-
Projected loss of brown macroalgae and seagrasses with global environmental change
Nature Communications (2024)
-
Cryptic diversity in southern African kelp
Scientific Reports (2024)
-
Carbon export from seaweed forests to deep ocean sinks
Nature Geoscience (2024)
-
Past climate-driven range shifts structuring intraspecific biodiversity levels of the giant kelp (Macrocystis pyrifera) at global scales
Scientific Reports (2023)