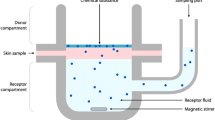
Abstract
Skin permeation is an essential biological property of small organic compounds our body is exposed to, such as drugs in topic formulations, cosmetics, and environmental toxins. Despite the limited availability of experimental data, there is a lack of systematic analysis and structure. We present a novel resource on skin permeation data that collects all measurements available in the literature and systematically structures experimental conditions. Besides the skin permeation value kp, it includes experimental protocols such as skin source site, skin layer used, preparation technique, storage conditions, as well as test conditions such as temperature, pH as well as the type of donor and acceptor solution. It is important to include these parameters in the assessment of the skin permeation data. In addition, we provide an analysis of physicochemical properties and chemical space coverage, laying the basis for applicability domain determination of insights drawn from the collected data points. The database is freely accessible under https://huskindb.drug-design.de or https://doi.org/10.7303/syn21998881.
Measurement(s) | skin permeation of chemicals • skin permeation |
Technology Type(s) | digital curation |
Factor Type(s) | chemical compound • skin source site • skin layer • skin preparation technique • temperature • storage duration • pH • acceptor medium • donor medium |
Sample Characteristic - Organism | Homo sapiens |
Machine-accessible metadata file describing the reported data: https://doi.org/10.6084/m9.figshare.13172831
Similar content being viewed by others
Background & Summary
Skin is the biggest organ in the human body. It provides a physical barrier to external influences, contributes to the metabolising and excretory functions and can absorb chemical substances. Skin has a layered structure, and it is assumed that a chemical compound, which permeates all layers of epidermis, the uppermost layer of the skin, is being fully carried away into the bloodstream by the capillaries of the dermis1. The transport itself is driven by a concentration gradient, and the classical model of permeation describes three absorption pathways: intracellular, intercellular and follicular. Investigation of skin permeation properties of chemical compounds is crucial for occupational exposure risk assessment and cosmetic and pharmaceutical development1. For this reason, skin permeation by xenobiotics has been investigated for many years2,3,4,5,6,7,8,9. Despite the time and cost intensiveness of the necessary experiments to determine skin permeation properties, the amount of data in the public domain has been steadily growing. This enabled research groups to collect enough data to develop predictive mathematical models. Different modelling approaches were implemented using a multitude of algorithms and data handling procedures. For example, Bratt et al.10, used permeation values from the Flynn dataset11 and removed values, that did not fit the overall data, leaving 60 data points. Chen et al.12 used permeation data on 215 compounds as a target value and did not report any outlier handling. Degim et al.13 used data on 40 substances and did not describe any outlier handling steps. A publication by Tsakovska et al.14 gives a comprehensive overview of these and other record collections.
A careful and thorough analysis of all these approaches revealed that most existing datasets such as the ones compiled by Flynn et al.11 or Wilschut et al.15 do not contain the experimental conditions under which the skin permeability measurements were obtained. Others, like the once created by Vecchia et al.16 or Magnusson et al.17 include experimental parameters. One of the most comprehensive data collections is the EDETOX database18 which returns 537 data points when querying in vitro measurements done with human skin. Not all of these data points, however, contain the skin permeation value (or steady-state flux and concentration of the chemical which can be used to calculate skin permeation). Apart from that, EDETOX does not contain the information on the skin source: whether the skin was obtained post-mortem from a cadaver or during a cosmetical procedure. No database containing all experimental conditions and skin permeation records exists to date. Additionally, when carefully comparing and analysing published datasets, inconsistencies can be identified, such as different or simply copied values. For example, Patel et al. used a logkp value for 2-ethoxyethanol of −7.16 (cm/s). This value was also used by Baba et al.19, Neely et al. (OSU-KP)20, Chen et al.12 and Lian et al.21. In contrast, Brown et al.22 used a logkp value of −6.92 (cm/s). For corticosterone Wilschut et al.15 and Patel et al.23 both used a logkp value of −7.78 (cm/s), but Baba et al.19 and Lian et al.21 used a value of −7.08 (cm/s), whereas Lian et al. included additional values of −7.56 (cm/s) and −6.81 (cm/s). In addition to this, different research groups used different measuring units. Wilschut et al. used kp measured in cm/h, Patel et al. logkp in cm/h and Lian et. al. logkp in cm/s. It is also important to consider that different layers of epidermis have different properties in regard to skin permeation: the hydrophobic stratum corneum is a barrier for hydrophilic substances. The hydrophilic viable epidermis, which is positioned under the stratum corneum, represents a barrier to hydrophobic substances. Thus, it is important to consider, which layer of the skin was used to obtain experimental data when analysing skin permeation values.
In this study, we present a novel comprehensive database of skin permeation values and corresponding experimental protocols. This manually curated database is a result of an extensive and careful analysis of previously published human skin permeation datasets and publications containing skin permeation data available in the public domain which additionally includes associated experimental protocols wherever available.
Methods
We performed a rigorous search and analysis of the original publications that reported skin permeability values to create a comprehensive, systematic and structured database.
Data content and organisation
Inclusion criteria for the data from a publication were as follows:
-
publication was accessible in the public domain
-
publication was the primary source of the data
-
the kp value was reported in the paper or it was possible to calculate it from the given steady-state flux and concentration by division (for the sake of data uniformity, huskinDB contains logarithmic kp values measured in cm/s)
-
data was discarded if no quantitative kp value was reported and it was obtainable only from the plots in the publication
-
permeation was measured using undamaged human skin and aqueous solution (in some cases with the addition of ethanol or methanol or in organic solvent) or neat compound.
If the publication satisfied the criteria mentioned above, the following experimental parameters were taken from it in addition to the kp value:
-
name of the compound
-
whether the skin was obtained during a surgical procedure or from a cadaver
-
site of the body from which skin was excised
-
skin preparation technique
-
layer of the skin
-
storage temperature
-
storage duration
-
whether the compound was tested neat or as a solution
-
temperature of the donor solution
-
pH of the donor solution
-
type of the donor solution
-
temperature of the acceptor solution
-
pH of the acceptor solution
-
type of the acceptor solution
-
type of the permeation cell
-
steady-state flux and concentration of the compound (if used for skin permeation calculation)
If parameters were unknown, the corresponding fields were left empty. Apart from the compound name, huskinDB contains canonical SMILES that were generated using Open Babel version 2.4.1 (http://openbabel.org)24.
For the purpose of data analysis, molecular weights and octanol/water partition coefficients (logP) were calculated using the RDKit version 2019.09.3 (https://github.com/rdkit/rdkit) library for Python (http://www.python.org).
Data Records
Conducted work has led to the development of the “Human Skin DataBase- huskinDB” (huskinDB25, https://doi.org/10.7303/syn21998881). As of April 2020, huskinDB25 contains 546 kp values for 251 different compounds covering 94 publications. This data can be accessed also via huskinDB homepage (https://huskindb.drug-design.de). On the huskinDB website, the user has full access to the records in the “Database” section and can perform searches. It is possible to define a custom set of rules to filter the data. If a specific compound is selected, all data on this compound is shown on a separate page. In the “Search” section of the huskinDB website, it is possible to draw a custom molecule using Marvin JS (https://chemaxon.com/products/marvin-js) and search for ten best matching compounds in the database (see Fig. 1). The content of the tables can be downloaded in the form of a CSV-file. Online-only Table 1 gives an overview of the experimental parameters and associated values that describe conditions under which skin permeation values were obtained. These parameters are discussed in greater detail below and correspond to the structure and the content of the downloadable CSV files.
Compound name
The database contains one name per compound. Only one name was chosen if a different name for the same compound was used in different publications. As shown in Online-only Table 1, the most frequently tested compound was testosterone with 16 data points. The second most frequent were estradiol and water with 15 data points each.
Skin donor type
Almost half of the skin permeation measurements were assessed using cadaver skin. The rest of the skin was obtained as a result of surgical procedures. The records also specify the type of surgical procedure. If the publication describes using skin from different sources, all of them are given. For example, Anderson et al.26 describe using skin obtained from a cadaver or during abdominoplasty. Therefore, the corresponding record is “Abdominoplasty or cadaver”.
Body site (skin source)
Excised abdominal skin was used most frequently, obtained either as a result of abdominoplasty, e.g. Legoabe et al.27, or removed from a cadaver, e.g. Blank et al.28. Different sites are listed if different sources were reported in the publication, e.g. Johnson et al.29 used skin that was removed from the chest, back and abdomen.
Skin preparation technique
Heat separation was the most frequent method used to prepare the skin. With this technique, previously obtained skin tissue is heated in water at 60°C for one minute, which leads to a separation of the epidermis30. The second most frequently reported technique was cutting the skin in different thicknesses using a dermatome - an instrument that cuts the upper layer of the skin of defined thickness31. If the thickness of the dermatomed skin was reported, it was noted in the database. If the authors used dermatomed skin but did not report the obtained thickness, no thickness is given, for example, Kushla et al.32 used dermatomed skin of thickness 150μm and the corresponding entry “dermatomed 150μm” can be found in the in huskinDB, Boogaard et al.33 did not specify the thickness of dermatomed skin and as such the corresponding entry reads “dermatomed”.
Layer of the skin used
Analysis of the original publications revealed that research groups used different skin layers to obtain permeability values. As described in the introduction, skin layers have different properties with respect to their absorption of chemicals. This has been recognised by many researchers26,34,35,36. Therefore, it was important for us to include this parameter in the database. If the layer was reported in the original publication, it was directly transferred to huskinDB. Otherwise, the type of the layer was determined from the skin preparation technique, and the body site as follows: if heat separation was used, the corresponding entry is “epidermis”30, if the authors wrote that the skin was “cut and placed”, “fat removed” or “dermatomed” without specifying the dermatome thickness, the corresponding entry is “epidermis, dermis”. Otherwise, information from Table 1 was used if the authors reported the thickness of the dermatomed skin and the skin source site. Analysis of the collected data shows that the most frequently used layer was epidermis, followed by epidermis with dermis. This can be explained through the use of heat separation (most frequently used preparation technique) and dermatome (second most frequently used preparation technique) respectively.
The Flux and Notes columns give optional additional information. The first contains steady-state flux and concentration values which were used to calculate skin permeation if it was not directly reported in the publication (by division of the flux value by the concentration). The second one gives clarification on data point obtainment or processing if it was deviating from the overall data point obtainment protocol, e.g. if the kp measurements was obtained using a suspension as a donor medium.
Other parameters were directly transferred from the original publications.
huskinDB25 database is licensed under a Creative Commons Attribution 4.0 International License.
Technical Validation
The following steps were performed in order to confirm that the records in huskinDB are correctly transferred from the original publications: first, the collected skin permeation values were compared to the ones reported in other data sets if these data sets contained skin permeation values from the same publication. After that, the correctness and completeness of all records was controlled by double-checking the content of the publications which reported experimental data. On this stage of validation errors in approximately 4% of records were identified and corrected. Next, after the initial compilation of huskinDB, all data records were double-checked twice by two different scientists. This validation did not reveal any errors in the data. Subsequently, a random collection of data points (approximately 10%) has been chosen and double-checked once again. No errors were identified on this stage as well.
Usage Notes
One of the aims of creating the huskinDB was to facilitate the development of in silico methods for predicting skin permeation or to gain additional knowledge of processes behind skin permeation. If developed, such predictive models or insights need to specify their applicability domain, which describes the chemical space in which valid predictions can be performed or to which some hypothesis is applicable37,38,39. This applicability domain directly depends on the database that is used to create predictive models. During an analysis of the previously created models for skin permeation, molecular weight and octanol/water partition coefficient were identified as typical descriptor variables13,23,40,41,42. These two descriptors were also found to improve the accuracy of logkp predicting models that were developed in-house. Therefore, molecular weight and octanol/water partition coefficient were chosen to analyse the applicability domain of huskinDB. Additionally, skin permeation value (logkp) was used to evaluate the chemical space since applicability domain is bound to its target variable. Below, a brief description of the chemical space of a hypothetical model or hypothesis, that is acquired using the full huskinDB dataset is provided. Obtained skin permeation values range between −11.436 (cm/s) (Propranolol hydrochloride) and −1.778 (cm/s) (1-Nonalol) with a mean value of −6.36 (cm/s) (SD = 1.36, SE = 0.059). This is illustrated in Fig. 2(a), where logkp values in the histogram form a skewed Gaussian distribution. Digitoxin (764.4) and disodium octaborate tetrahydrate (590.0) show the largest molecular weight, whilst water (18.0) and methanol (32.0) form the lower molecular weight limit. A histogram of molecular weights in Fig. 2(b) shows a bias towards compounds with lower values. A plot of molecular weight against logkp in Fig. 2(c) reveals that the coverage of the chemical space is non-homogeneous.
For chemical space analysis, 8,697 molecules were obtained from the DrugBank version 5.0.1143 (37 molecules were omitted: it was either not possible to process them with RDKit, or their molecular weight was larger than 2,500 or their calculated logP was higher than 20 or less than −20). The distribution of data from DrugBank and huskinDB over molecular weight and octanol/water partition coefficient (calculated with RDKit) in Fig. 3 illustrates that compounds from huskinDB only represent a subset of the DrugBank molecules: they show a smaller spread on both axes and represent molecules with relatively small weights.
Additional information on experimental conditions in huskinDB can be used to explain phenomena that were not correctly interpreted or understood before. In the following example, this use case is shown on the skin permeation values of three steroid subsets that were found by research groups to be inconsistent with other measurements: 1995, Barratt et al.10 identified 12 measurements of hydrocortisone derivatives in the Flynn dataset that were described as outliers. All of these measurements were performed by the same research group - Anderson et al.26 (further referred to as Anderson steroids). Later, Abraham et al.44 reported that the permeation values of steroids measured by Scheuplein et al.5 (Scheuplein steroids) are too low in comparison to more recent values published by Johnson et al.29 (Johnson steroids). To assess the correctness of both steroid data subsets, Abraham et al.44 developed a model that was not based on any of these values and evaluated its predictions. Abraham et al.44 showed that predictions are in better agreement with Johnson’s data. Another interesting observation was made by Abraham et al., who suggested that Anderson steroids have kp values that are too high.
Figure 4 shows the relationship between logkp values and the molecular weight of three steroid subsets. The permeation values of different groups are clustered; however, when analysed more closely, it is apparent that the data points are grouped based on the molecular weights. Further data analysis reveals that these values were obtained using different skin layers. Permeation of Scheuplein and Johnson steroids were measured using epidermis, whereas Anderson steroids were measured using thinner layer- stratum corneum. This means that the difference in the skin permeability between the steroid subsets might be caused by the difference in molecular weights on the one hand and by different skin layers on the other.
Steroids measured by Scheuplein, Johnson and Anderson are grouped in clusters. It is apparent that these researchers used compounds with different molecular weight classes and performed experiments with different skin layers; in green: Scheuplein steroids, in blue: Johnson steroids, in red: Anderson steroids; the layer of the skin that was used is shown by the shape of the marker – square: epidermis, diamond: stratum corneum. Scheuplein steroids, in blue: Johnson steroids, in red: Anderson steroids; the layer of the skin that was used is shown by the shape of the marker – square: epidermis, diamond: stratum corneum.
This demonstrates that the application area of huskinDB goes beyond permeation value prediction. It can also be used to assess the influence of experimental variables on the kp value. However, it must be noted that huskinDB contains molecules with small molecular weights in comparison to databases such as DrugBank43. Apart from that, data on experimental protocols of many measurements is unknown. Therefore, a direct comparison of the data points is not always possible. For example, misleading conclusions can be drawn when comparing two measurements with one obtained using aqueous solution and another using unknown donor medium, which may be non-aqueous. Careful analysis of the skin membrane with respect to the lipophilicity of the chemical compound is advised as well when looking at a specific skin permeation measurement. A skin membrane consisting solely of stratum corneum will provide a weaker barrier function than a membrane consisting of the whole epidermis and dermis, especially for the lipophilic compounds since the hydrophilic viable epidermis may become the rate-determining factor.
Also, we would like to point out that the user might not be interested in the kp value itself, rather in the flux values across the skin. These values might provide a descriptor more adequate for estimation of the compound intake during, for example, occupational exposure and can be calculated by multiplication of the kp value by the compound concentration in the donor medium. Given that the compound solubility is used as the concentration, one might calculate the theoretical maximum flux. The solubility, in its turn, is dependent on the medium in which the compound is dissolved. This also denotes the importance of the data point quality validation since the donor medium and thus the solubility is different across the data records in huskinDB, making direct kp value comparison rather challenging and, in some cases, even misleading. Different properties of different solvents also mean that the kp value of some compound obtained using one solvent, cannot be used to predict the flux value of the same compound in another solvent. Therefore, it is vital for the user to perform a proper data handling and to account for the limited chemical space and unknown values prior creating mathematical models or drawing conclusions from the data. In this regard, defining a custom set of rules would rank the data point suitability for one’s domain of interest in a tailored way. For example, a measurement obtained using a donor solution containing 90% ethanol or a measurement with many unknown parameters might be ranked as a “low confidence record” and excluded from further processing. In contrast, a data point obtained using common and known parameters such as phosphate-buffered saline as donor and acceptor medium at 37 °C and heat-separated epidermis would be ranked as a “high confidence record” and included in subsequent analysis. It is also important to note that more reliable data on compounds from different chemical classes would further expand the applicability domain of huskinDB. We plan to update the database, and therefore researchers are encouraged to submit their data to huskinDB, which provides a technical framework for the structured deposition of skin permeability data for small organic molecules.
Code availability
The code which was used to create Figs. 2–4 and analyse the data records in huskinDB can be found under the following link: https://github.com/RhDm/huskinDB_publication This repository contains a detailed guide on how to install the requirements and run the code.
References
Bolzinger, M.-A., Briançon, S., Pelletier, J. & Chevalier, Y. Penetration of drugs through skin, a complex rate-controlling membrane. Curr Opin Colloid & Interface Sci 17, 156–165 (2012).
Overton, E. Ueber die allgemeinen osmotischen Eigenschaften der Zelle, ihre vermutlichen Ursachen und ihre Bedeutung für die. Physiologie. Vierteljahresschr D Naturforsch Ges in Zürich 44, 88–135 (1899).
Url, W. Charles Ernest Overton, 75 Jahre Lipoidtheorie. Verh Zool Bot Ges Wien 115, 24–33 (1976).
Scheuplein, R. J. Mechanism of percutaneous adsorption. I. Routes of penetration and the influence of solubility. J Invest Dermatol 45, 334–346 (1965).
Scheuplein, R. J., Blank, I. H., Brauner, G. J. & MacFarlane, D. J. Percutaneous absorption of steroids. J Invest Dermatol 52, 63–70 (1969).
Scheuplein, R. J. & Blank, I. H. Permeability of the skin. Physiol Rev 51, 702–747 (1971).
Scheuplein, R. J. & Blank, I. H. Mechanism of percutaneous absorption. IV. Penetration of nonelectrolytes (alcohols) from aqueous solutions and from pure liquids. J Invest Dermatol 60, 286–296 (1973).
Blank, I. H., Scheuplein, R. J. & MacFarlane, D. J. Mechanism of percutaneous absorption. 3. The effect of temperature on the transport of non-electrolytes across the skin. J Invest Dermatol 49, 582–589 (1967).
Michaels, A. S., Chandrasekaran, S. K. & Shaw, J. E. Drug permeation through human skin: theory andinvitro experimental measurement. AIChE Journal 21, 985–996 (1975).
Barratt, M. D. Quantitative structure-activity relationships for skin permeability. Toxicol In Vitro 9, 27–37 (1995).
Flynn, G. L. In Principles of Route-to-Route Extrapolation for Risk Assessment (ed Gerrity, T. R. & Henry C. J.) 93–127 (Elsevier, 1990).
Chen, L. J., Lian, G. P. & Han, L. J. Prediction of human skin permeability using artificial neural network (ANN) modeling. Acta Pharmacol Sin 28, 591–600 (2007).
Degim, T., Hadgraft, J., Lbasimis, S. & Özikan, Y. Prediction of skin penetration using artificial neural network (ANN) modeling. J Pharm. Sci 92, 656–664 (2003).
Tsakovska, I. et al. Quantitative structure-skin permeability relationships. Toxicology 387, 27–42 (2017).
Wilschut, A., ten Berge, W. F., Robinson, P. J. & McKone, T. E. Estimating skin permeation. The validation of five mathematical skin permeation models. Chemosphere 30, 1275–1296 (1995).
Vecchia, B. E., Bunge, A. L. In Transdermal Drug Delivery Systems (ed Hadgraft, J.) Ch. 3 (CRC Press, 2002).
Magnusson, B. M., Anissimov, Y. G., Cross, S. E. & Roberts, M. S. Molecular size as the main determinant of solute maximum flux across the skin. J Invest Dermatol 122, 993–999 (2004).
Williams, F. M. EDETOX. Evaluations and predictions of dermal absorption of toxic chemicals. Int Arch Occup Environ Health 77, 150–151 (2004).
Baba, H., Takahara, J. & Mamitsuka, H. In silico predictions of human skin permeability using nonlinear quantitative structure-property relationship models. Pharm Res 32, 2360–2371 (2015).
Neely, B. J., Madihally, S. V., Robinson, R. L. Jr. & Gasem, K. A. Nonlinear quantitative structure-property relationship modeling of skin permeation coefficient. J Pharm Sci 98, 4069–4084 (2009).
Lian, G., Chen, L. & Han, L. An evaluation of mathematical models for predicting skin permeability. J Pharm Sci 97, 584–598 (2008).
Brown, T. N., Armitage, J. M., Egeghy, P., Kircanski, I. & Arnot, J. A. Dermal permeation data and models for the prioritization and screening-level exposure assessment of organic chemicals. Environ Int 94, 424–435 (2016).
Patel, H., ten Berge, W. & Cronin, M. T. Quantitative structure-activity relationships (QSARs) for the prediction of skin permeation of exogenous chemicals. Chemosphere 48, 603–613 (2002).
O’Boyle, N. M. et al. Open Babel: An open chemical toolbox. J Cheminform 3, 33 (2011).
Stepanov, D. huskinDB. Synapse https://doi.org/10.7303/syn21998881 (2020).
Anderson, B. D., Higuchi, W. I. & Raykar, P. V. Heterogeneity effects on permeability-partition coefficient relationships in human stratum corneum. Pharm Res 5, 566–573 (1988).
Legoabe, L. J., Breytenbach, J. C., N’Da, D. D. & Breytenbach, J. W. In-vitro transdermal penetration of cytarabine and its N4-alkylamide derivatives. J Pharm Pharmacol 62, 756–761 (2010).
Blank, I. H. Penetration of low-molecular-weight alcohols into skin. I. Effect of concentration of alcohol and type of vehicle. J Invest Dermatol 43, 415–420 (1964).
Johnson, M. E., Blankschtein, D. & Langer, R. Permeation of steroids through human skin. J Pharm Sci 84, 1144–1146 (1995).
Kassis, V. & Sondergaard, J. Heat-separation of normal human skin for epidermal and dermal prostaglandin analysis. Arch Dermatol Res 273, 301–306 (1982).
Tamer, S. In Skin Grafts - Indications, Applications and Current Research (ed Spear, M.) Ch. 1 (IntechOpen, 2011).
Kushla, G. P. & Zatz, J. L. Influence of pH on lidocaine penetration through human and hairless mouse skin in vitro. Int. J. Pharm 71, 167–173 (1991).
Boogaard, P. J., Denneman, M. A. & Van Sittert, N. J. Dermal penetration and metabolism of five glycidyl ethers in human, rat and mouse skin. Xenobiotica 30, 469–483 (2000).
Wilkinson, S. C. et al. Interactions of skin thickness and physicochemical properties of test compounds in percutaneous penetration studies. Int Arch Occup Environ Health 79, 405–413 (2006).
Henning, A., Neumann, D., Kostka, K. H., Lehr, C. M. & Schaefer, U. F. Influence of human skin specimens consisting of different skin layers on the result of in vitro permeation experiments. Skin Pharmacol Physiol 21, 81–88 (2008).
Lee, F. W., Earl, L. & Williams, F. M. Interindividual variability in the percutaneous penetration of testosterone through human skin in vitro. Toxicology 168, 63–63 (2001).
Sahigara, F. et al. Comparison of different approaches to define the applicability domain of QSAR models. Molecules 17, 4791–4810 (2012).
Dimitrov, S. et al. A stepwise approach for defining the applicability domain of SAR and QSAR models. J Chem Inf Model 45, 839–849 (2005).
European Union. Regulation (EC) No 1907/2006 of the European parliament and of the council of 18 December 2006 concerning the registration, evaluation, authorisation and restriction of chemicals (REACH), establishing a European chemicals agency, amending directive 1999/45/EC and repealing council regulation (EEC) No 793/93 and commission regulation (EC) No 1488/94 as well as council directive 76/769/EEC and commission directives 91/155/EEC, 93/67/EEC, 93/105/EC and 2000/21/EC. Off. J. Eur. Union 49, L 396 (2006).
Potts, R. O. & Guy, R. H. Predicting skin permeability. Pharm Res 9, 663–669 (1992).
Cronin, M. T., Dearden, J. C., Moss, G. P. & Murray-Dickson, G. Investigation of the mechanism of flux across human skin in vitro by quantitative structure-permeability relationships. Eur J Pharm Sci 7, 325–330 (1999).
Neumann, D., Kohlbacher, O., Merkwirth, C. & Lengauer, T. A fully computational model for predicting percutaneous drug absorption. J Chem Inf Model 46, 424–429 (2006).
Wishart, D. S. et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res 46, D1074–D1082 (2018).
Abraham, M. H., Martins, F. & Mitchell, R. C. Algorithms for skin permeability using hydrogen bond descriptors: the problem of steroids. J. Pharm. Pharmacol 49, 858–865 (1997).
Sandby-Moller, J., Poulsen, T. & Wulf, H. C. Epidermal thickness at different body sites: relationship to age, gender, pigmentation, blood content, skin type and smoking habits. Acta Derm Venereol 83, 410–413 (2003).
Robertson, K. & Rees, J. L. Variation in epidermal morphology in human skin at different body sites as measured by reflectance confocal microscopy. Acta Derm Venereol 90, 368–373 (2010).
Niu, Y. et al. Reduced dermis thickness and AGE accumulation in diabetic abdominal skin. Int J Low Extrem Wounds 11, 224–230 (2012).
Chan, J. C., Ward, J., Quondamatteo, F., Dockery, P. & Kelly, J. L. Skin thickness of the anterior, anteromedial, and anterolateral thigh: a cadaveric study for split-skin graft donor sites. Arch Plast Surg 41, 673–678 (2014).
Olenius, M. & Johansson, O. Variations in epidermal thickness in expanded human breast skin. Scand J Plast Reconstr Surg Hand Surg 29, 15–20 (1995).
Acknowledgements
We thank Lhasa Limited for funding and supporting the project. We are also immensely grateful to Martyn Chilton, Pearl Saville and Scott McDonald for comments that improved the manuscript. Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
D.S. and G.W. conceived the study. D.S. collected the data, and developed the website. D.S., G.W. and S.C. wrote the manuscript. G.W. and S.C. supervised the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Online-only Table
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
The Creative Commons Public Domain Dedication waiver http://creativecommons.org/publicdomain/zero/1.0/ applies to the metadata files associated with this article.
About this article
Cite this article
Stepanov, D., Canipa, S. & Wolber, G. HuskinDB, a database for skin permeation of xenobiotics. Sci Data 7, 426 (2020). https://doi.org/10.1038/s41597-020-00764-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-020-00764-z
This article is cited by
-
An update of skin permeability data based on a systematic review of recent research
Scientific Data (2024)
-
Fragment contribution models for predicting skin permeability using HuskinDB
Scientific Data (2023)
-
Predicting skin permeability using HuskinDB
Scientific Data (2022)
-
A curated binary pattern multitarget dataset of focused ATP-binding cassette transporter inhibitors
Scientific Data (2022)
-
Studying and mitigating the effects of data drifts on ML model performance at the example of chemical toxicity data
Scientific Reports (2022)