Abstract
The Protein Ontology (PRO) provides an ontological representation of protein-related entities, ranging from protein families to proteoforms to complexes. Protein Ontology Linked Open Data (LOD) exposes, shares, and connects knowledge about protein-related entities on the Semantic Web using Resource Description Framework (RDF), thus enabling integration with other Linked Open Data for biological knowledge discovery. For example, proteins (or variants thereof) can be retrieved on the basis of specific disease associations. As a community resource, we strive to follow the Findability, Accessibility, Interoperability, and Reusability (FAIR) principles, disseminate regular updates of our data, support multiple methods for accessing, querying and downloading data in various formats, and provide documentation both for scientists and programmers. PRO Linked Open Data can be browsed via faceted browser interface and queried using SPARQL via YASGUI. RDF data dumps are also available for download. Additionally, we developed RESTful APIs to support programmatic data access. We also provide W3C HCLS specification compliant metadata description for our data. The PRO Linked Open Data is available at https://lod.proconsortium.org/.
Similar content being viewed by others
Introduction
As an evolving extension to the current hypertext document web, Linked Open Data (LOD) is a new paradigm where data are published and interconnected on the web using open standards such as Uniform Resource Identifiers (URIs), Hypertext Transfer Protocol (HTTP), Resource Description Framework (RDF), Web Ontology Language (OWL) and SPARQL Protocol and RDF Query Language (SPARQL). This enables data from heterogeneous sources to be shared, integrated and queried in a web of data. Tim Berners-Lee in his web architecture note introduced a set of best practices for publishing and interlinking structured data on the web, also well known as the Linked Data principles1: (1) Use URIs as names of things; (2) Use HTTP URIs, so that people can look up those names; (3) When someone looks up a URI, provide useful information using the standards (RDF, SPARQL etc.); and (4) Include links to other URIs, so that they can discover more things.
Bio2RDF2 is an open source project that uses Semantic Web technologies to build a large network of Linked Open Data for Life Sciences from a diverse set of heterogeneously formatted sources obtained from multiple data providers. It uses federated SPARQL queries to facilitate continuous integration of life sciences data from resources such as Mouse Genome Informatics (MGI)3, Saccharomyces Genome Database (SGD)4, Rat Genome Database (RGD)5, NCBI’s Gene resources (NCBIGene)6, HUGO Gene Nomenclature Committee (HGNC)7, and Nematode Information Resource (WormBase)8. The European Bioinformatics Institute (EBI) provides an RDF platform9 that facilitates answering complex research questions through queries/exploration of integrated resources, including UniProtKB10, Reactome11, Ensembl12, and Gene Ontology13.
As formal and explicit specifications of domains of interest, ontologies consist of terms representing precisely defined entities and the relationships between them. Ontologies are increasingly being used to define the basic terms and relations in biological domains, often as the foundation for search, integration and exchange of biological data. The Protein Ontology (PRO)14 provides an ontological representation of protein-related entities, notably, including those of organism-neutral nature15. In addition to the ontology itself, PRO includes annotations and cross-reference information, and can be used to facilitate the discoverability and aggregation of data related to protein entities in the context of protein functions, pathways and drug targets. The Protein Ontology has been used by the research community for applications including named entity recognition/tagging16,17,18, entity definition (for example, of cell types19), and as an import for protein-related terms in other ontologies20,21. PRO Linked Open Data and SPARQL endpoint have been used to assist in orthology mapping22 and to study kinase post-translational modifications and cancer-associated mutations23. The PRO website has garnered more than 20 million hits in the past year. PRO Linked Open Data and SPARQL endpoint alone garnered 321,000 hits. PRO is ranked among the top 5 endpoints by YummyData SPARQL endpoint monitor24.
In this paper we introduce the PRO RDF data models and the metadata description of PRO Linked Open Data. We provide faceted browser, SPARQL endpoint with graphical user interface (GUI), and downloadable RDF files. We demonstrate federated SPARQL queries to answer questions for biological knowledge discovery using multiple connected resources.
Results
Each PRO term represents a distinct class of organism-neutral or organism-specific protein entities (e.g., modified forms, orthologous isoforms, and protein complexes). To help readers understand PRO RDF data models, we present an exemplar PRO term in each category using RDF Graph, RDF/XML, and Turtle formats. Links for these are provided in the relevant section, however an example RDF Graph of one PRO term (PR:000046294, RAC-alpha serine/threonine-protein kinase phosphorylated 3 (human)) is also shown in Fig. 1 and listed as Turtle format below.
@prefix owl:<http://www.w3.org/2002/07/owl#>.
@prefix rdfs:<http://www.w3.org/2000/01/rdf-schema#>.
@prefix xsd:<http://www.w3.org/2001/XMLSchema#>.
@prefix obo:<http://purl.obolibrary.org/obo/>.
@prefix oboInOwl:<http://www.geneontology.org/formats/oboInOwl#>.
obo:PR_000046294 a owl:Class;
rdfs:comment “Category = organism-modification.”;
rdfs:label “RAC-alpha serine/threonine-protein kinase phosphorylated 3 (human)”;
rdfs:subClassOf obo:PR_000049939, obo:PR_P31749;
rdfs:subClassOf [a owl:Restriction;
owl:onProperty obo:RO_0002160;
owl:someValuesFrom obo:NCBITaxon_9606
];
rdfs:subClassOf [a owl:Restriction;
owl:onProperty obo:BFO_0000051;
owl:someValuesFrom obo:MOD_00046
];
obo:IAO_0000115 “A RAC-alpha serine/threonine-protein kinase phosphorylated 3 in human. UniProtKB:P31749, Ser-473, MOD:00046.”;
oboInOwl:hasDbXref “Reactome:R-HSA-377265”;
oboInOwl:hasExactSynonym “UniProtKB:P31749, Ser-473, MOD:00046”, “hAKT1/Phos:3”;
oboInOwl:hasOBONamespace “protein”;
oboInOwl:id “PR:000046294”;
owl:equivalentClass [a owl:Class;
owl:intersectionOf (obo:PR_000049939
[a owl:Restriction;
owl:onProperty obo:RO_0002160;
owl:someValuesFrom obo:NCBITaxon_9606])].
]
)
].
obo:RO_0002160 rdfs:label “only_in_taxon”.
obo:NCBITaxon_9606 rdfs:label “Homo sapiens”.
obo:PR_000049939 oboInOwl:hasExactSynonym “AKT1/Phos:3”.
obo:PR_P31749 oboInOwl:hasExactSynonym “hAKT1”.
obo:BFO_0000051 rdfs:label “has_part”.
obo:MOD_00046 rdfs:label “O-phospho-L-serine”.
PRO uniform resource identifiers (URIs)
PRO follows the rule of reusing existing identifiers whenever feasible, e.g., http://purl.obolibrary.org/obo/GO_0032991 (Gene ontology), http://purl.obolibrary.org/obo/RO_0002160” (Relations Ontology), etc. The format of PRO identifier is “PR_xxxxxxxxx”, e.g., http://purl.obolibrary.org/obo/PR_000000001. However, for UniProtKB related entries incorporated into PRO organism-gene or organism-sequence categories, we use the UniProtKB accession number and prefix it with “PR_” as its corresponding PRO identifier. e.g., http://purl.obolibrary.org/obo/PR_P00015.
Types of PRO terms
Protein Ontology terms are organized into general categories, often with organism-specific versions. Below we describe these categories. Example terms for each category are given in Table 1. Whenever possible, PRO attempts to make connections between orthologs. Extending from the concept of orthology between genes, we have introduced the concept of orthologous proteoforms. For example, ortho-isoforms are isoforms that are believed to have arisen prior to speciation and divergence of the primary sequence. That is, ortho-isoforms were true alternative isoforms (as defined above) in a common ancestor and are, quite likely, functionally equivalent. Ortho-modified forms are modified versions of ortho-isoforms where the modification(s) occur on equivalent residues.
Category = family
Each PRO term at the family level refers to protein products of a distinct gene family arising from a common ancestor. The leaf-most nodes at this level are usually families comprising paralogous sets of gene products (of a single or multiple organisms). For example, smad2 and smad3 both encode proteins that are TGF receptor-regulated while smad1, smad5, and smad9 are all BMP receptor-regulated. Thus, “TGF-beta receptor-regulated smad protein” and “BMP receptor-regulated smad protein” are terms denoting distinct families. Note that this level collectively refers to any such grouping at any level of similarity. For example, the two families indicated above can merge into a “receptor-regulated smad protein” class and further merge (with the protein products of smad4, smad6, and smad7) into the “smad protein” class.
Category = gene
Each PRO term at the gene level refers to the protein products of a distinct gene in a reference organism and the orthologs thereof. For example, “smad2” and “smad3” are two different genes, and therefore have two different PRO entries at the gene level of distinction. The protein products of what is recognized as smad2 in humans and what is recognized as smad2 in mouse fall under the single gene-level term “smad2”. Thus, a single term at the gene-level distinction collects the protein products of (usually 1-to-1) orthologs for that gene. Organism-specific versions are (typically) defined logically as the intersection of the parent (gene-level) and the organism (taxonomic) terms. If a resource provides gene information, PRO will indicate this using the has_gene_template relation.
Category = seqgroup
Each PRO term at the seqgroup level refers to proteins encoded by the same gene that are distinguished from siblings based on differences in shared portions of encoding mRNAs. That is, all members of a given seqgroup are encoded by mRNAs that have a common subset of sequence features, either as a common subset of exons or as a common subset of sequence variations. Examples include proteins encoded by the PTPRC (CD45) gene, where each member of the CD45R subtype (CD45RA, CD45RAB, CD45RAC) minimally contains exon 4 (aka ‘A’) even while each member of a given subtype has other exons that make them distinct, and histocompatibility genes such as HLA-A, where each member of a given subtype (HLA-A*24, HLA-A*68, etc) shares a common set of variations even while each member of a given subtype has other variations that make them distinct.
Category = sequence
Each PRO term at the sequence level refers to the protein products that arise from different alleles of a given gene (sequence variants), from splice variants of a given RNA, or from alternative initiation and ribosomal frameshifting during translation. One can think of this as a mature mRNA-level distinction. For example, smad2 encodes both a long splice form and a short splice form. The protein products of each isoform are separate PRO terms. If there is clear knowledge on the equivalency of isoforms (that is, they are “ortho-isoforms”)14, then the equivalent terms from different organisms are defined as children of the sequence level terms. For example, ARF GTPase-activating protein GIT1 isoform 1 from mouse (PR:Q68FF6-1), ARF GTPase-activating protein GIT1 isoform 1 from rat (PR:Q9Z272-1), and ARF GTPase-activating protein GIT1 isoform 3 from human (PR:Q9Y2X7-3) are ortho-isoforms that are children of the sequence level term PR:000044155. If the equivalency of isoforms is not well-established, the organism-sequence term is defined as a child of the organism-gene level term.
Category = modification
Each PRO term at the modification level refers to the protein products derived from a single mRNA species that differ because of some change (or lack thereof) that occurs after the initiation of translation (co- and post-translational). This includes sequence differences due to cleavage and chemical changes to one or more amino acid residues. In general, PRO does not provide time or space information, i.e., the order in which modifications occur or the location in which they are found unless such are required to properly define the term. PRO only represents the final modified objects. If there is clear knowledge on the equivalency of the modified forms from different organisms, then the equivalent organism-modification terms are defined as children of the modification level terms. For example, RAC-alpha serine/threonine-protein kinase phosphorylated 3 from human (PR:000046294) and mouse (PR:000049940) are ortho-modified forms that are phosphorylated on the position equivalent to Ser-473 of the human protein. In the PRO hierarchy, they are children of the modification level term PR:000049939. If the equivalency of the modified forms is not well-established, the organism-modification term is defined as a child of the appropriate organism-gene term.
Category = complex
Each PRO term at the complex level25 ultimately traces to the general term “protein-containing complex” found in GO (GO:0032991), defined as “A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which at least one component is a protein and the constituent parts function together”. Indeed, complex-level terms in PRO are imported from GO whenever possible. However, because GO complexes are (predominantly) defined with respect to function, PRO will create complex terms when a component-specific definition is desired. When doing so, PRO represents complexes explicitly–that is, by subunit composition–defining each member of the complex at the level of its isoform, variant, or modified form, whenever possible. Most complex-related terms defined by PRO are organism-specific. Note that a protein in its monomeric state, linked non-covalently to a small chemical, is not considered a protein complex by the above definition, but it can be defined as a subclass of the protein term using the CHEBI ID for the small chemical and the relation “non-covalently_bound_to”.
Dataset description
A high-quality consistent metadata description is essential to the successful discovery, exchange, and query of a Linked Dataset. The Protein Ontology Linked Open Data is accompanied with metadata description using Vocabulary of Interlinked Datasets (VoID), the Provenance vocabulary (PROV) and Dublin Core vocabulary, which are compliant with the W3C HCLS specification26. The metadata for Protein Ontology Linked Open Data is described at three levels: (1) summary level, which provides a description of a dataset that is independent of a specific version or format; (2) version level, which captures version-specific characteristics of a dataset; and (3) distribution level, which captures metadata about a specific form and version of a dataset.
Linkset is a way of identifying the content that links instances in one dataset with instances in another dataset. A separate linkset is created for each link predicate relating a particular pair of datasets. A linkset is a subset of the dataset which publishes it. The linkset itself is of type void:Linkset and provides the same metadata as a RDF distribution. The statistics relevant for a linkset are the number of triples it contains and are reported using the void:triples property. The full VoID description of PRO Linked Open Data is at https://lod.proconsortium.org/releases/latest/void.ttl.
Federated SPARQL query
Federated SPARQL query is a powerful method that allows the simultaneous search of multiple data resources and aggregates the results from multiple SPARQL endpoints. PRO RDF data can be combined with other RDF data, such as UniProt10 RDF data (at https://sparql.uniprot.org/) and DisGeNET27 RDF data (at http://rdf.disgenet.org/sparql/) as demonstrated below to generate new biological insights that are not possible using one individual resource.
Sample federated SPARQL query 1
To answer the question, “Which human proteins are potentially involved in disease via loss of function”, the query is designed to get all human proteins in PRO whose UniProtKB counterpart has variants with loss of function implicated in disease. The partial query result is shown in Fig. 2 as a knowledge graph.
Query 1:
PREFIX obo:<http://www.geneontology.org/formats/oboInOwl#>
PREFIX up:<http://purl.uniprot.org/core/>
PREFIX ud:<http://purl.uniprot.org/database/>
PREFIX up:<http://purl.uniprot.org/core/>
PREFIX taxon:<http://purl.uniprot.org/taxonomy/>
PREFIX rdf:<http://www.w3.org/1999/02/22-rdf-syntax-ns#>
PREFIX rdfs:<http://www.w3.org/2000/01/rdf-schema#>
PREFIX faldo:<http://biohackathon.org/resource/faldo#>
SELECT DISTINCT?
PRO_term
(STR(?_PRO_label) AS?PRO_label)
(STRAFTER(STRBEFORE(STR(?_PRO_category), “.”), “ = “) AS?PRO_category)
?Protein
?Variant
(STR(?_Text) AS?Description)
WHERE
{
SERVICE<http://sparql.uniprot.org/sparql>
{
?Protein rdfs:seeAlso?PRO_term.?
PRO_term
up:database ud:PRO;
a up:Resource.?Protein a up:Protein.?Protein up:organism taxon:9606.?Protein up:annotation?Variant.?Variant a up:Natural_Variant_Annotation.?Variant rdfs:comment?_Text.
FILTER (CONTAINS(?_Text, ‘loss of function’))
}
?PRO_term rdfs:label?_PRO_label.
?PRO_term rdfs:comment?_PRO_category.
}
Sample federated SPARQL query 2
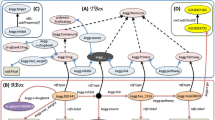
To answer the question, “What disease(s) are associated with AlzForum-derived sequence variants described in PRO”, we construct a query to find variants in UniProt or DisGeNET for AlzForum (https://www.alzforum.org/) PRO terms. The partial query result is shown in Fig. 3 as a knowledge graph.
Knowledge graph of exemplary query result of federated SPARQL query 2 (Find variants in UniProt or DisGeNET for AlzForum PRO terms). Ellipse and circle shapes are RDF nodes. Rectangle shapes are RDF literals. Directed edges are RDF properties. Circle shapes represent anonymous classes or blank nodes.
Query 2:
PREFIX obo:<http://purl.obolibrary.org/obo/>
PREFIX up:<http://purl.uniprot.org/core/>
PREFIX ud:<http://purl.uniprot.org/database/>
PREFIX taxon:<http://purl.uniprot.org/taxonomy/>
PREFIX rdf:<http://www.w3.org/1999/02/22-rdf-syntax-ns#>
PREFIX rdfs:<http://www.w3.org/2000/01/rdf-schema#>
PREFIX sio:<http://semanticscience.org/resource/>
PREFIX ncit:<http://ncicb.nci.nih.gov/xml/owl/EVS/Thesaurus.owl#>
PREFIX dcterms:<http://purl.org/dc/terms/>
PREFIX so:<http://purl.obolibrary.org/obo/SO_>
PREFIX oboInOwl:<http://www.geneontology.org/formats/oboInOwl#>
PREFIX skos:<http://www.w3.org/2004/02/skos/core#>
SELECT DISTINCT
?PRO
(?PRO_Label AS?Name)
(STRAFTER(STRBEFORE(STR(?PRO_Category), “.”), “ = “) AS?Category)
(STRBEFORE(STRAFTER(STR(?PRO_Category), “.”), “.”) AS?Comment)
(?protein AS?UniProt)
(?uniprotvar AS?UniProt_Var)
(?variant AS?DisGeNET_Var)
(STR(?variantTitle) AS?RS_ID)
(STR(?vscore) AS?VDAScore)
(?disease AS?Disease)
(STR(?diseaselabel) AS?DiseaseName)
(?gene AS?Gene)
(STR(?gSymbol) AS?GeneSymbol)
(STR(?geneTitle) AS?GeneName)
WHERE {
{
SELECT?PRO?Parent?PRO_Label?PRO_Category?uniprotvar
WHERE {
VALUES?PRO {obo:PR_000037702 obo:PR_000037753}
?PRO rdfs:subClassOf?Parent.
?Parent rdfs:seeAlso?uniprotvar.
?PRO rdfs:label?PRO_Label.
?PRO rdfs:comment?PRO_Category.
FILTER(CONTAINS(LCASE(?PRO_Category), “by = alzforum”))
FILTER(CONTAINS(LCASE(STR(?uniprotvar)), “annotation”))
}
}
SERVICE<http://sparql.uniprot.org/sparql>
{
?uniprotvar
a up:Natural_Variant_Annotation;
rdfs:seeAlso?_rs;
skos:related?up_disease.
?up_disease
a up:Disease;
rdfs:comment?up_disease_comment.
FILTER(CONTAINS(LCASE(?up_disease_comment), “alzheimer”))
[]
rdf:object?uniprotvar;
rdf:predicate up:annotation;
rdf:subject?protein;
a rdf:Statement.
BIND(IRI(REPLACE(STR(?_rs), “purl.uniprot.org”, “identifiers.org”)) AS?variant)
}
SERVICE<http://rdf.disgenet.org/sparql/>
{
OPTIONAL {
?vda sio:SIO_000628?variant,?disease;
sio:SIO_000216?vscoreIRI.?vscoreIRI sio:SIO_000300?vscore.?variant dcterms:title?variantTitle.?disease rdfs:label?diseaselabel.?disease dcterms:title?diseaseTitle.
FILTER(CONTAINS(LCASE(?diseaseTitle), “alzheimer”))
OPTIONAL {?variant so:associated_with?gene.?gene a ncit:C16612.?gene dcterms:title?geneTitle.?gene sio:SIO_000205?symbolUri.?symbolUri dcterms:title?gSymbol.
}
}
}
} ORDER BY DESC(?vscore)
Discussion
The guiding principles of FAIR data aim to facilitate the discovery, integration, and analysis of relevant datasets by humans and machines by making them Findable (F), Accessible (A), Interoperable (I), and Reusable (R)28. As a community resource, we strive to follow FAIR principles, disseminate regular updates of our data, support multiple methods for accessing, querying and downloading data in various formats, and provide documentation both for scientists and programmers. The formal FAIRness assessment29,30 results for Protein Ontology Linked Open Data are presented in Table 2. According to the results, PRO follows the FAIR principles quite well. Moreover, according to the YummyData SPARQL endpoint monitor, PRO (https://yummydata.org/endpoint/129) ranks among the top 5 endpoints by the Umaka Score24 calculated based on six criteria: Availability, Freshness, Operation, Usefulness, Validity and Performance.
Nonetheless, there are still some improvements to be made, and these will be addressed in the future work. For example, we plan to improve PRO entry pages to support content negotiation with structured data, such as JSON-LD to provide explicit clues about the meaning of the PRO term to search engines.
PRO has a user-friendly web interface that supports search, browse and retrieval of PRO terms and related information at https://proconsortium.org/. The PRO website also provides a query interface to the SPARQL endpoint, and new functionality using Linked Open Data is being developed. Web statistics show an increasing proportion of accesses to the SPARQL query interface over text searches, indicating the trends towards using the SPARQL server and programmatic access.
Methods
Data imported into PRO comes largely from well-known curated sources (for example, UniProtKB and Reactome) that are updated with each release through a combination of manual and automated means. Special attention is given to terms for which positional information is cited. Upon import, and regularly thereafter, such positions are verified against the current protein sequence (for example, to ensure that a named amino acid is at the indicated position on the sequence). Data integrity and consistency of ontology is checked prior to each release using the ELK reasoner31, a very fast reasoner that supports the EL subset of OWL 2 as part of the ROBOT tool32.
Users can query PRO Linked Open Data using SPARQL at https://sparql.proconsortium.org/virtuoso/sparql, which is powered by OpenLink Virtuoso server community edition (http://vos.openlinksw.com/owiki/wiki/VOS) (version 07.20.3217) with the faceted browser, SPARQL 1.1 query federation and Cross-Origin Resource Sharing (CORS) enabled to support a range of complex and federated queries that merge data from other SPARQL endpoints.
Faceted browser
Protein Ontology Linked Open Dataset can be accessed via Virtuoso Faceted Browser (Fig. 4), a general-purpose RDF data query facility for data exploration by faceted browsing over entity relationship types (i.e. relations).
SPARQL GUI
Protein Ontology Linked Open Data can be accessed via YASGUI (Yet Another Sparql GUI), a web application to query any SPARQL endpoint. YASGUI provides various advanced features for creating, sharing, and visualizing SPARQL queries and their results. We also provided a comprehensive set of example SPARQL queries (Fig. 5).
RESTful APIs
As a full query language, SPARQL can be difficult for some people to learn. We therefore developed RESTful APIs (Fig. 6) for programmatic access to Protein Ontology Linked Open Data for data integration or analysis. The API specification was designed using the Swagger™ Editor based on OpenAPI Specification 3. Swagger UI was used to visualize and interact with the API’s resources automatically generated from API specifications. The PRO APIs include 8 API operation groups and 34 access paths as shown in Table 3 and are implemented using the Django-REST framework.
Users can use the API web interface to interactively customize the API requests. The API web pages also provide users with dynamically generated source code (client libraries) for common scripting and programming languages that provide examples with their query options and filters fully integrated that can be used as standalone scripts or programs or integrated into users’ large projects.
RDF dumps
RDF dumps of Protein Ontology Linked Open Data are available for download in application/rdf + xml or text/turtle formats with corresponding RDF centric statistics, as shown in Table 4. Each dataset is loaded into a named graph in the Virtuoso triple store. A set of six RDF centric metrics are computed for each dataset that summarize their contents:
-
total number of triples
-
total number of distinct classes (the number of distinct resources occurring as objects of rdf:type)
-
total number of entities
-
total number of distinct subject nodes
-
total number of distinct predicates
-
total number of distinct object nodes
Data availability
RDF dumps can be downloaded at https://lod.proconsortium.org/release.html. Virtuoso faceted browser can be accessed at https://sparql.proconsortium.org/virtuoso/fct/. RESTful APIs can be accessed at https://lod.proconsortium.org/api.html. SPARQL GUI can be accessed at https://lod.proconsortium.org/yasgui.html. The formal FAIRness assessment results can be accessed at https://lod.proconsortium.org/fair.html.
Code availability
The source code that implements https://lod.proconsortium.org/ website is available from GitHub at https://github.com/PROconsortium/PRoteinOntology/tree/master/pro_lod
The source code that implements PRO RESTful APIs is available from GitHub at https://github.com/PROconsortium/PRoteinOntology/tree/master/proapi
References
Berners-Lee, T. Linked Data, https://www.w3.org/DesignIssues/LinkedData.html (2006).
Callahan, A. et al. Bio2RDF release 2: improved coverage, interoperability and provenance of life science linked data. In: Cimiano P., Corcho O., Presutti V., Hollink L., Rudolph S. (eds) The Semantic Web: Semantics and Big Data. ESWC 2013. Lecture Notes in Computer Science. 7882, 200-212 (Springer, Berlin, Heidelberg, 2013).
Bult, J. et al. Mouse genome database (MGD). Nucleic Acids Res. 47, D801–D806 (2019).
Cherry, M. et al. Saccharomyces genome database: the genomics resource of budding yeast. Nucleic Acids Res. 40, D700–705 (2012).
Smith, R. et al. The year of the rat: the rat genome database at 20: a multi-species knowledgebase and analysis platform. Nucleic Acids Res. 48, D731–D742 (2020).
National Center for Biotechnology Information. Gene https://www.ncbi.nlm.nih.gov/gene/ (2020).
Yates, B. et al. Genenames.org: the hgnc and vgnc resources in 2017. Nucleic Acids Res. 45, D619–D625 (2017).
Todd, W. et al. WormBase: a modern model organism information resource. Nucleic Acids Res. 48, D762–D767 (2020).
Jupp, S. et al. The ebi rdf platform: linked open data for the life sciences. Bioinformatics 30, 1338–1339 (2014).
The UniProt Consortium. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 47, D506–D515 (2019).
Jassal, B. et al. The reactome pathway knowledgebase. Nucleic Acids Res. 48, D498–D503 (2020).
Yates, A. et al. Ensembl 2020. Nucleic Acids Res. 48, D682–D688 (2020).
The Gene Ontology Consortium. The gene ontology resource: 20 years and still going strong. Nucleic Acids Res. 47, D330–D338 (2019).
Natale, D. et al. Protein ontology (PRO): enhancing and scaling up the representation of protein entities. Nucleic Acids Res. 45, D339–D346 (2017).
Neal, L. et al. Harmonizing semantic annotations for computational models in biology. Brief Bioinform. 20, 540–550 (2019).
Wang, D. et al. AAgAtlas 1.0: a human autoantigen database. Nucleic Acids Res. 45, D769–D776 (2017).
Bada, M. et al. Concept annotation in the craft corpus. BMC Bioinformatics 13, 161 (2012).
Pyysalo, S. et al. LION lbd: a literature-based discovery system for cancer biology. Bioinformatics 35, 1553–1561 (2019).
Overton, A. et al. Reporting and connecting cell type names and gating definitions through ontologies. BMC Bioinformatics 20, 182 (2019).
He, Y. et al. OSCI: standardized stem cell ontology representation and use cases for stem cell investigation. BMC Bioinformatics 20, 180 (2019).
Lin, Y., Xiang, Z. & He, Y. Ontology-based representation and analysis of host-brucella interactions. J. Biomed. Semantics 6, 37 (2015).
Zeinab, F. et al. Murine oviductosomes (ovs) microrna profiling during the estrous cycle: delivery of ovs-borne micrornas to sperm where mir-34c-5p localizes at the centrosome. Sci. Rep. 8, 16094 (2018).
Huang, L. et al. Integrative annotation and knowledge discovery of kinase post-translational modifications and cancer-associated mutations through federated protein ontologies and resources. Sci. Rep. 8, 6518 (2018).
Yamamoto, Y., Yamaguchi, A. & Splendiani, A. YummyData: providing high-quality open life science data. Database 2018, bay022 (2018).
Bult, C. et al. The representation of protein complexes in the protein ontology (pro). BMC Bioinformatics 12, 371 (2011).
W3C Interest Group. Dataset descriptions: hcls community profile. http://www.w3.org/TR/2015/NOTE-hcls-dataset-20150514/ (2015).
Piñero, J. et al. The disgenet knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 48, D845–D855 (2020).
Wilkinson, M. et al. The fair guiding principles for scientific data management and stewardship. Sci. Data 3, 160018 (2016).
Wilkinson, M. et al. A design framework and exemplar metrics for fairness. Sci. Data 5, 180118 (2018).
Garcia, L. et al. FAIR adoption, assessment and challenges at UniProt. Sci. Data 6, 175 (2019).
Kazakov, Y., Krötzsch, M. & Simančík, F. The Incredible elk - from polynomial procedures to efficient reasoning with el ontologies. Journal of Automated Reasoning 1, 1–61 (2014).
Jackson, R. et al. ROBOT: a tool for automating ontology workflows. BMC Bioinformatics 1, 407 (2019).
Acknowledgements
The authors thank our collaborators from Protein Ontology Consortium. The funding is provided by the National Institute of General Medical Sciences [R01GM080646].
Author information
Authors and Affiliations
Contributions
Author contributions are as follows: Writing [C.C., H.H., K.R., J.C., C.A., C.W. and D.N.]; curation [K.R., D.N. and C.A.]; programming [C.C., H.H. and J.C.]; discussion [C.C., H.H., K.R., J.C., C.A., C.W. and D.N.].
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chen, C., Huang, H., Ross, K.E. et al. Protein ontology on the semantic web for knowledge discovery. Sci Data 7, 337 (2020). https://doi.org/10.1038/s41597-020-00679-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-020-00679-9
This article is cited by
-
RTX-KG2: a system for building a semantically standardized knowledge graph for translational biomedicine
BMC Bioinformatics (2022)