Abstract
Measures of chlorophyll represent the algal biomass in freshwater lakes that is often used by managers as a proxy for water quality and lake productivity. However, chlorophyll concentrations in lakes are dependent on many interacting factors, including nutrient inputs, mixing regime, lake depth, climate, and anthropogenic activities within the watershed. Therefore, integrating a broad scale dataset of lake physical, chemical, and biological characteristics can help elucidate the response of freshwater ecosystems to global change. We synthesized a database of measured chlorophyll a (chla) values, associated water chemistry variables, and lake morphometric characteristics for 11,959 freshwater lakes distributed across 72 countries. Data were collected based on a systematic review examining 3322 published manuscripts that measured lake chla, and we supplemented these data with online repositories such as The Knowledge Network for Biocomplexity, Dryad, and Pangaea. This publicly available database can be used to improve our understanding of how chlorophyll levels respond to global environmental change and provide baseline comparisons for environmental managers responsible for maintaining water quality in lakes.
Measurement(s) | chlorophyll a • phosphorus atom • nitrogen atom • dissolved carbon atom in water • dissolved oxygen in water • volume • lake surface area • depth of water • pH |
Technology Type(s) | digital curation |
Factor Type(s) | geographic location |
Sample Characteristic - Environment | freshwater lake |
Sample Characteristic - Location | Earth (planet) |
Machine-accessible metadata file describing the reported data: https://doi.org/10.6084/m9.figshare.12928352
Similar content being viewed by others
Background & Summary
Lake water accounts for less than 1% of the world’s surface freshwater supply, but provides critical ecosystem services, including consumption, transportation, agriculture, and recreation, in addition to habitat for over 100,000 species of invertebrates, insects, animals, and plants1,2,3. However, freshwater lakes are vulnerable to the effects of water fouling, nutrient enrichment, and alterations in climate and land use owing to their sensitivity to local and global environmental changes4,5,6. Alterations in biological and chemical lake processes can affect how and when freshwater resources can be used. Particularly, increases in lake chlorophyll levels can impact water quality through alterations in colour and odor7, dissolved oxygen availability8, and overall lake production9.
Chlorophyll a (chla) is frequently used as a straightforward and suitable representative measurement of lake productivity and water quality10,11,12. Many environmental assessments commonly use chla as a biological indicator for determining lake trophic status10,13. In freshwater ecology, chla also functions as a good proxy for other biological variables, such as primary production, and is often included as a covariate in limnological studies14. Chla is therefore routinely measured in water quality programs across the globe, making a good candidate for the focus of a water quality database with broad spatial coverage.
To discern the limnological processes that determine chla in lakes requires consideration of water chemistry, lake morphometry, and the landscape setting. While lakes naturally vary in their chla concentrations owing to seasonal fluctuations and climate variability, they can also respond to anthropogenic influences such as nutrient inputs15. Anthropogenic sources of nutrient loadings in lakes include runoff from the surrounding watershed from land use changes16,17, atmospheric deposition18, and sewage discharge19. Furthermore, individual lake properties such as surface area, depth, and volume can mediate the temperature, productivity, and energy flow of a lake20. Accordingly, water chemistry (defined here as total phosphorus, total nitrogen, dissolved organic carbon, and dissolved oxygen) as well as numerous morphometric characteristics were included in the assembly of this database.
There are two main methods for generating chla data, either from model-inferred estimates using remotely sensed images or through in situ sampling. There are chla levels inferred from remote sensing21,22 that can be effective for comparisons among lakes, but these are less common because there is significant error surrounding the separation of turbidity from light attenuation in the water column23,24. Similarly, the extent of in situ measurements can be restricted because certain lakes are difficult to access (e.g. high alpine, or arctic). Ideally, a chla database would have both modelled and field measurements to allow users the option to trade-off spatial coverage for accuracy.
Building on recent extensive national water quality databases25, we fill a strong need for a cohesive and broad-scale database of water quality worldwide. The incentive to assemble this database of lake chla, water chemistry, and morphometric characteristics was to identify chla patterns over broad spatial and temporal scales. Other applications of this database include and are not limited to identifying which environmental stressors (e.g. climate, nutrient or anthropogenic factors) are most important in driving changes in water quality, specifically chla. Using the published scientific literature and online data repositories, we conducted a systematic review to acquire instances where chla has been measured. Here, we present a database of wide spatial coverage of chla from 11,959 lakes distributed across 72 countries collected in situ or by satellites. From these same data sources, we also acquired information about lake morphometry and water chemistry as they are highly correlated with chla concentrations. We provide a summary of these data and associated variables to serve as a tool in ecological research and freshwater management.
Methods
Data acquisition
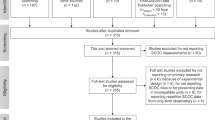
We obtained data by conducting a systematic review of the literature and searching for published repositories in online databases. We first conducted a systematic review to identify relevant primary articles using “chlorophyll” and “lake*” as citation search terms in Web of Science between the years 2000 and 2018. From these published manuscripts, we acquired chla and other water chemistry data for 11,959 lakes worldwide. Papers that were not primary articles or were not in a field relating to limnology were excluded. We screened 3322 articles published between 2000 and 2018 because this timeframe represents more recent lake conditions (e.g. post zebra mussel invasion in North America) with minimal repetition. We excluded articles if the methods used to collect water quality data violated the following criteria: i) were not sampled in the lake (i.e. from a sediment core); ii) were collected in a manipulative study (i.e. from a mesocosm or other experimental modification of the lake’s water chemistry); or iii) were monitored in situ using sensors that were not supplemented by additional calibration techniques26 (Fig. 1). All lakes also required reporting of latitude and longitude. If an article did not violate any criteria, we extracted data from tables, in text, or through the digitization of figures using WebPlotDigitizer (https://apps.automeris.io/wpd/) either from the article or from the supplementary data (Fig. 1). Extracted data included values for chla, total phosphorus, total nitrogen, dissolved organic carbon, and dissolved oxygen. If data were unavailable from the manuscript, we contacted the study authors to request their data (Fig. 1). We also collected data on lake volume, surface area, mean depth, maximum depth, Secchi depth, and pH when available within the study.
We found an additional 15 online data repositories that contained lake chla measurements and other water chemistry data for 10,997 lakes using the online search engines Dryad (https://datadryad.org/), The Knowledge Network for Biocomplexity (KnB - https://knb.ecoinformatics.org/), Github (https://github.com/) and Google Dataset Search (https://toolbox.google.com/datasetsearch) also using the search terms “chlorophyll” and “lake*”. Information on each of the repositories can be found in Table 1. The data repositories were predominantly from the USA. The repository with the greatest number of observations and lakes was, by far, repo14 (92% of all observations and 69% of all lakes) that was a previous data compilation effort by Soranno et al.25 across multiple government agencies and research partners (Table 1). The methods varied for observations within this database but are well documented by the authors25. The sources of data obtained from these repositories were diverse, coming from government programs, independent research groups, Long-Term Ecological Research sites, and non-profit monitoring agencies.
Chlorophyll data
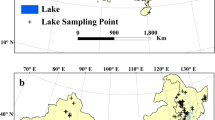
Our team acquired chla data for 228,168 unique survey instances in 11,959 lakes distributed across 72 countries and on every continent including Antarctica (Fig. 2). In all but 47 instances, data were measured in situ. In 10 datasets, chlorophyll was estimated using remote sensed data from satellite imagery. Although remote sensed data can be less precise than in situ surveys, we included this data because it provides estimates in lakes that are difficult to access. There were 37 instances where it was unclear which methodologies were used that are identified in the methodsData.csv. Each chla measurement was converted to standardized units (mg L−1) and corresponds with the lake’s latitude, longitude, and the year in which the measurement was taken (Table 2). In some cases, the same lake was sampled in multiple locations (which were associated with different coordinates within the lake’s perimeter) and/or sampled multiple times within the same year (e.g. monthly; which was associated with a sampling date and not just the year). Almost all datasets used surface measurements (41.4%) or an integrated water sample (23.8%). Only 7.7% of collected datasets used a specific depth for their measurements (the remaining were undescribed). The deepest sample collected was 250 m below the surface from Lake Baikal27. The detection limits for studies were often 0.1 μg L−1 or lower (71% of collected datasets), although some were coarse including 6% of collected datasets that had detection limits at 100 μg L−1. We flagged observations where the detection limits were greater than the observed value of chla (<1.6% of observations) because these values may be inaccurate and should be treated with caution. For instance, values of zero are likely not true zeros but may represent chla measurements below the detectable limits of the method used. There were 454 observations (0.12%) that had zero values.
Sampling method techniques varied including analysis by spectrophotometry, fluorometry, or the methods by Eaton and Franson28. A comprehensive discussion of the details of each of the standard methods of chla extraction can typically be found in individual manuscripts. Generally, water samples were passed through a filter, and then chla was extracted from the organism atop the filter using an organic solvent (e.g. acetone or ethanol). The chla concentrations were then determined by a spectrophotometer (to record light absorbance of chla at a specific wavelength) or by a fluorometer (to record light fluorescence of chla at a specific wavelength). Other methods of data collection included high performance liquid chromatography and sonication/freeze-thaw method.
The chlorophyll values reported were often aggregated values that were collected across multiple profiles of the water column, different points within the same lake (spatial), or the same location but over multiple times (temporal). We documented the replication within each of the collected datasets when provided for each of these three categories (column, spatial, and temporal) in the methodsData.csv (Table 3). We included both a qualitative and quantitative description of replication in these three categories from each individual dataset. The measurement type would either be described as a raw value, meaning it was collected and reported from a single observation, or it would be reported as an aggregate statistic (i.e. mean or median). The NumObs column within the methodsData.csv represents the number of values that were extracted from the respective study or online repository. This number was typically smaller than the Replicate column which represented the number of observations collected by the original data contributors of the study or repository. The Replicate column can often be estimated by multiplying the number of replicates in the column profile, the areas sampled within the lake (spatial), and the number of times it was sampled (temporal). However, there were cases where the Replicate column was not divisible by these three categories because of uneven sampling. For example, one lake may have been sampled at three different depths but another sampled at only one. When multiple depths were provided, we calculated an average for each water chemistry variables to create an integrated water sample and provide the details of the depths surveyed in the methodsData.csv. In all other cases (i.e. temporal and spatial), we maintained each replicate within a lake as a separate observation in our dataset when the authors provided this information. There was considerable variability in the number of replicates that were collected between studies and repositories. For instance, repo10 collected values every five minutes (n = 254,527) that were collated into annual averages over four years for two lakes (n = 8). By contrast other lakes were sampled considerably less, such as a series of Patagonian and Pampean lakes that were sampled once annually for two years29. We provide details of all available data on replication in the methodsData.csv file to allow for accurate comparisons between studies.
Water chemistry and geomorphometric data
We compiled total phosphorus (TP; mg L−1), total nitrogen (TN; mg L−1), dissolved organic carbon (DOC; mg L−1), and dissolved oxygen (DO; mg L−1) measurements from sampling observations which also presented, at a minimum, lake chla data, sampling date and geographic coordinates (Tables 2; 4). The methodology used to obtain in situ water chemistry data varied among studies and is described in the methodsData.csv. Generally, water chemistry samples were analyzed spectrophotometrically, fluorometrically, or by a multi-parameter water quality probe (e.g. Yellow Springs Instrument, which was supplemented with additional calibration methods to ensure measurement accuracy). We also collected lake volume, surface area, mean depth, maximum depth, Secchi depth, and pH from the original data provider when available within the study (Tables 2; 4). Secchi depth was often measured using either a 30 cm white circular disk or a slightly modified 20 cm disk with black and white patterning. Almost all studies used portable pH meters to measure water pH.
Unique identification
We assigned a unique identifier (hereafter survey instance, labeled “uniqueID”) in the dataset to every chla data point separated by unique lake, GPS coordinate, year, month, and study. We could not treat every spatial coordinate as an independent lake because some coordinates were surveyed within the same lake either within or among studies. To determine unique lake identifiers that correspond with each survey instance, we used the HydroLAKES database of lake location and shape30 (http://wp.geog.mcgill.ca/hydrolab/hydrolakes/). We matched the spatial polygons of lakes present within the HydroLAKES database with the spatial coordinates extracted from the studies. In instances where the survey instance did not match a lake within HydroLAKES database, we conducted a Google search to determine if the lake was unique from others. Using these methods, we generated a unique lake identifier associated with each of our survey instances. The country was determined from the geographic coordinates of the lake.
Data Records
We have published the MS_citations and Repo_citations in an open access repository31 (Filazzola et al. 2020. Knowledge Network for Biocomplexity. https://doi.org/10.5063/F1RV0M1S) with data from the published manuscripts and data repositories (Table 1) that were systematically processed to extract chla data (Table 3). Each of these files contains citation information such as the authors, year that the study was published, location published (e.g. journal, data repository), and whether the dataset was ultimately included within this database (Table 3). Each of these files lists studies that were explored as potentially having chla data but were excluded.
The main dataset file ChlaData.csv contains general information about each survey instance that connects across the other files by the uniqueID identifier (Table 3). The first column has a unique identifier that corresponds with every survey instance that is separated by year, month, geospatial point, and study. This file is to be used for subsetting the survey points for respective analyses, such as within a certain timeframe or country. This file also contains a column of lake identifiers corresponding to each of the survey instances because, within and among studies, some lakes were surveyed multiple times at different locations. All water chemistry variables reported, including chla are reported in mg L−1 (Table 4). Finally, the dataset includes information about the morphometric lake characteristics when reported, such as surface area, mean lake depth, and maximum lake depth (Table 4).
Technical Validation
We conducted quality control and quality assurance across the database to validate the data from each of the independent sources. In total, there were 228,168 unique survey instances that required quality assurance and quality control (QA/QC). We separated our QA/QC into three distinct stages: 1) Import and Compilation, 2) Unique Lake Identification, and 3) Value Validation and Conversion.
Import and compilation
Data extraction from each study was conducted by separate individuals and contributed to a master data file. After the data were assembled from each of the manuscripts and online repositories, we conducted initial examination of transcription errors such as variables placed in the wrong columns, variables missing units, or incorrect characters. To ensure there were no duplicates in data across the multiple datasets we examined samples collected in the same year, that were within 1 km of each other, and had chla values within 0.00005 mg L−1 of each other. A random subset of 10% from our database was validated by members within our group that were different than the original person that collected the data. These spot checks were meant to mitigate errors that could have been generated when compiling the database or converting values to the same units.
Unique lake identification
We checked the coordinates for each survey instance to ensure it represented an actual body of water not in the ocean. When possible, the name of the lake described in the paper was compared to the described coordinates. Any errors or erroneous observations in location (e.g. negative longitudes for studies in the eastern hemisphere) were determined by comparing study descriptions with points and using a map of the lakes (Fig. 2). We compared maps of lakes within 1 km of each other but with different lake identifiers to ensure these were indeed separate lakes. To determine if any lakes were incorrectly identified as the same, we searched for any uniqueIDs that had the same lake identifier but were more than 100 km apart. In total, there were 1,374 lakes that we identified belonging to multiple studies such as Lake Taihu (20 studies), Lake Ontario (16 studies), and Lake Chao (8 studies).
Value validation
We compared the distribution of all values to identify potential outliers that could indicate an incorrect measurement. The units across all datasets were standardized to all be mg L−1, and were converted from multiple other units including µg L−1, mg m−3, and g m−3. All lakes that had units mg m−2 were removed because they were based on downscaling of surface water only and did not convert properly to mg L−1. We rounded all values of chla to 0.0001 mg L−1 (0.1 µg L−1) because analytical equipment used within studies rarely had better precision. A full list of the sensitivities from each dataset can be found within the methodsData.csv dataset.
We generated histograms and compared the distributions of each variable to identify observations that could be erroneous. For all water chemistry variables, we flagged any observation that was above three standard deviations from the mean. These values were then compared to the original data source to ensure it was correctly transcribed. We conducted these flagging exercises excluding repo14 because that specific repository was extremely large relative to the other observations, is biased towards north-east USA, and has been extensively validated previously25. We explored all values that exceeded (>1 mg L−1) for the water chemistry variables. Many of the extremely high values (>1 mg L−1) were from a study by Marselina and Burhanudin32 that measured the water quality of extremely polluted lakes in Indonesia. The highest value recorded for chla was 4.33 mg L−1 taken from Binder Lake, Iowa in 2006 during what we believe was an algal bloom. We explored a log-transformed distribution of chla values and found the median chla value across all observations was 6.0 µg L−1 (Fig. 3). Approximately 30% of the observations were considered oligotrophic with chla values less than 2.5 µg L−1 (Fig. 3). Lakes with chla values of zero were observed in some arctic and alpine lakes. We observed two noticeable differences in symmetry in the histogram of chla that could potentially be explained by the detection limits of some devices for chla (e.g. 1.0 µg L−1) or trophic boundaries (e.g. oligotrophic vs mesotrophic). We also explored the distribution of the other water chemistry and lake morphometry variables (Fig. 4). Using boxplots, we examined outliers that may have not been flagged from comparisons of extreme values. By comparing observations outside of the boxplots, we identified values that may have suffered from conversion errors as these would typically be off by three orders of magnitude (e.g. 1 mg L−1 = 1000 µg L−1). Boxplots also allowed exploration of the distribution, to determine if there is any skew in the data that could have been generated by incorrect units or compilation errors. Any observations that were flagged were checked by exploring the initial dataset from which the values were obtained.
Distribution of water chemistry and lake morphometry values from database. Values represent log-transformed equivalent of the units presented in Table 4, except pH which is already log-transformed.
Usage Notes
We provide code in R Version 3.5.133 within our guide to join all files by their unique identifier for further analysis. Data synthesis and technical validation was conducted using tidyr34, and dplyr35. Visual quality assurance and figures were generated using ggplot236. To compensate for some missing lake characteristics that were not reported in the searched manuscripts, such as lake volume, depth, or surface area, we suggest that authors use additional resources such as LakeNet (http://www.worldlakes.org/), Global Water Bodies database – GLOWABO37, International Lake Environment Committee Foundation – ILEC (http://www.ilec.or.jp/en/), Global Lakes & Reservoirs Repository – GLR (http://www.worldlake-db.com), NHDPlus Version 2 (https://www.horizon-systems.com/NHDPlus) or HydroLAKES (http://wp.geog.mcgill.ca/hydrolab/hydrolakes). The HydroLAKES database is particularly useful and provides additional geomorphic data for approximately 1.4 million lakes globally30 (https://www.hydrosheds.org/). For other lake characteristics, the Global Lake Area, Climate, and Population (GLCP) dataset has synthesized climate and human population densities for more than 1.4 million lakes globally4. These datasets can complement the chlorophyll database built here to explore factors that drive water quality in freshwater lakes.
Code availability
All code for analyses included within this manuscript as well as meta-data files (including unique identifiers, repository and manuscript data, lake characteristics, water chla and chemistry data, and water sample collection method) are provided in an open access repository38. Within the repository, we also provide code for unit conversion (e.g. µg L−1 to mg L−1), and extracting climate data from the Climatic Research Unit at the University of East Anglia (http://www.cru.uea.ac.uk/).
References
Beeton, A. M. Large freshwater lakes: present state, trends, and future. Environ Conserv. 29, 21–38 (2002).
Shiklomanov, I. A. Water in Crisis: A Guide to the World’s Fresh Water Resources (Oxford Univ. Press, 1993).
McMichael, A. J., Woodruff, R. E. & Hales, S. Climate change and human health: present and future risks. The Lancet 367, 859–869 (2006).
Meyer, M. F., Labou, S. G., Cramer, A. N., Brousil, M. R. & Luff, B. T. The global lake area, climate, and population dataset. Sci. Data 7, 1–12 (2020).
Wrona, F. J. et al. Climate change effects on aquatic biota, ecosystem structure and function. Ambio 35, 359–369 (2006).
Adrian, R. et al. Lakes as sentinels of climate change. Limnol. Oceanogr. 54, 2283–2297 (2009).
Nürnberg, G. K. & Shaw, M. Productivity of clear and humic lakes: nutrients, phytoplankton, bacteria. Hydrobiologia 382, 97–112 (1998).
Makri, S., Lami, A., Lods-Crozet, B. & Loizeau, J. L. Reconstruction of trophic state shifts over the past 90 years in a eutrophicated lake in western Switzerland, inferred from the sedimentary record of photosynthetic pigments. J. Paleolimnol. 61, 129–145 (2019).
Håkanson, L. & Boulion, V. V. Regularities in primary production, Secchi depth and fish yield and a new system to define trophic and humic state indices for lake ecosystems. Int. Rev. Hydrobiol. 86, 23–62 (2001).
Carlson, R. E. A trophic state index for lakes. Limnol. Oceanogr. 22, 361–369 (1977).
Sterner, R. W. In situ-measured primary production in Lake Superior. J. Great Lakes Res. 36, 139–149 (2010).
Li, X., Sha, J. & Wang, Z. L. Chlorophyll-a prediction of lakes with different water quality patterns in China based on hybrid neural networks. Water 9, 524 (2017).
Vollenweider, R. & Kerekes, J. Eutrophication of Waters: Monitoring, Assessment and Control. OECD (1982).
Bennion, D. H., Warner, D. M., Esselman, P. C., Hobson, B. & Kieft, B. A comparison of chlorophyll a values obtained from an autonomous underwater vehicle to satellite-based measures for Lake Michigan. J. Great Lakes Res. 45, 726–734 (2019).
Elser, J. J. et al. Global analysis of nitrogen and phosphorus limitation of primary producers in freshwater, marine and terrestrial ecosystems. Ecol. Lett. 10, 1135–1142 (2007).
Hall, R. I., Leavitt, P. R., Quinlan, R., Dixit, A. S. & Smol, J. P. Effects of agriculture, urbanization, and climate on water quality in the northern Great Plains. Limnol. Oceanogr. 44, 739–756 (1999).
Bennett, E. M., Carpenter, S. R. & Caraco, N. F. Human impact on erodable phosphorus and eutrophication: a global perspective: increasing accumulation of phosphorus in soil threatens rivers, lakes, and coastal oceans with eutrophication. AIBS Bulletin 51, 227–234 (2001).
Williamson, C. E., Dodds, W., Kratz, T. K. & Palmer, M. A. Lakes and streams as sentinels of environmental change in terrestrial and atmospheric processes. Front. Ecol. Environ. 6, 247–254 (2008).
Carpenter, S. R. et al. Nonpoint pollution of surface waters with phosphorus and nitrogen. Ecol. Appl. 8, 559–568 (1998).
Williamson, C. E., Saros, J. E., Vincent, W. F. & Smol, J. P. Lakes and reservoirs as sentinels, integrators, and regulators of climate change. Limnol. Oceanogr. 54, 2273–2282 (2009).
Li, L., Li, L., Shi, K., Li, Z. & Song, K. A semi-analytical algorithm for remote estimation of phycocyanin in inland waters. Sci. Total Environ. 435, 141–150 (2012).
Odermatt, D., Danne, O., Philipson, P. & Brockmann, C. Diversity II water quality parameters from ENVISAT (2002-2012): a new global information source for lakes. Earth Syst. Sci. Data 10, 1527–1549 (2018).
Palmer, S. C., Kutser, T. & Hunter, P. D. Remote sensing of inland waters: Challenges, progress and future directions. Remote Sens. Environ. 157, 1–8 (2015).
Salama, M. S. & Verhoef, W. Two-stream remote sensing model for water quality mapping: 2SeaColor. Remote Sens. Environ. 157, 111–122 (2015).
Soranno, P. A. et al. LAGOS-NE: A multi-scaled geospatial and temporal database of lake ecological context and water quality for thousands of U.S. lakes. Gigascience 6, 1–22 (2017).
Zeng, L. H. & Li, D. L. Development of in situ sensors for chlorophyll concentration measurement. J. Sens. 2015, 1–16 (2015).
Shimaraeva, S. V., Pislegina, E. V., Krashchuk, L. S., Shchapov, K. S. & Silow, E. A. Dynamics of chlorophyll a concentration in the South Baikal pelagic during the direct temperature stratification period. Inland Water Biol. 10, 59–63 (2017).
Eaton, A. D., & Franson, M. A. H. Standard Methods for the Examination of Water and Wastewater. American Public Health Association, American Water Works Association, Water Environment Federation, Washington, Denver, Alexandria (2005).
Torremorell, A., del Carmen Diéguez, M., Queimaliños, C., Izaguirre, I. & Zagarese, H. E. Phytoplankton limitation in Patagonian and Pampean shallow lakes: effect of phosphorus and light. Hydrobiologia 816, 91–105 (2018).
Messager, M. L., Lehner, B., Grill, G., Nedeva, I. & Schmitt, O. Estimating the volume and age of water stored in global lakes using a geo-statistical approach. Nat. Commun. 7, 13603 (2016).
Filazzola, A. et al. A global database of chlorophyll and water chemistry in freshwater lakes. KNB Data Repository https://doi.org/10.5063/F1RV0M1S (2020).
Marselina, M. & Burhanudin, M. Trophic status assessment of Saguling Reservoir, Upper Citarum Basin, Indonesia. Air, Soil and Water Res. 10, 1–8 (2017).
R Development Core Team. R: A language and environment for statistical computing (2019).
Wickham, H. & Henry, L. tidyr: Tidy Messy Data. R package version 1.0.0. https://CRAN.R-project.org/package=tidyr (2019).
Wickham, H., François, R., Henry, L., & Müller, K. dplyr: A Grammar of Data Manipulation. R package version 0.8.3. https://CRAN.R-project.org/package=dplyr (2019).
Wickham, H. ggplot2: Elegant Graphics for Data Analysis. (Springer-Verlag, New York, 2016).
Verpoorter, C., Kutser, T., Seekell, D. A. & Tranvik, L. J. A global inventory of lakes based on high‐resolution satellite imagery. Geophys. Res. Lett. 41, 6396–6402 (2014).
Filazzola, A. afilazzola/ChlorophyllDataPaper: Initial-Release. Zenodo https://doi.org/10.5281/zenodo.3968735 (2020).
Hampton, S. E. et al. Ecology under lake ice. Ecol. Lett. 20, 98–111 (2017).
Karatayev, Vadim, A. et al. Eutrophication and Dreissena invasion as drivers of biodiversity: A century of change in the mollusc community of Oneida Lake. PloS One 9 (2014).
Richardson, D. C. et al. Transparency, geomorphology and mixing regime explain variability in trends in lake temperature and stratification across northeastern North America (1975–2014). Water 9, 442 (2017).
Mantzouki, E. et al. The European Multi Lake Survey (EMLS) dataset of physical, chemical, algal pigments and cyanotoxin parameters 2015. Environmental Data Initiative (2018).
Pollard, AminaI., Hampton, StephanieE. & Leech, DinaM. The Promise and Potential of Continental‐Scale Limnology Using the US Environmental Protection Agency’s National Lakes. Assessment. Limnol.Oceanogr. Bull. 27, 36–41 (2018).
Burnett, L., Moorhead, D., Hawes, I. & Howard-Williams, C. Environmental factors associated with deep chlorophyll maxima in Dry Valley lakes, South Victoria Land, Antarctica. Arct. Antarct. Alp. Res. 38, 179–189 (2006).
Takamura, N. & Nakagawa, M. The densities of bacteria, picophytoplankton, heterotrophic nanoflagellates and ciliates in Lake Kasumigaura (Japan) monitored monthly since 1996. Ecol. Res. 27, 839 (2012).
Gries, C., Gahler, M. R., Hanson, P. C., Kratz, T. K. & Stanley, E. H. Information management at the North Temperate Lakes Long-term Ecological Research site—Successful support of research in a large, diverse, and long running project. Ecol. Inform. 36, 201–208 (2016).
Acknowledgements
We thank all authors of the data that were used in generating this database for making their data available in an online repository or providing it through email. Without their extensive fieldwork, this database would not have been possible. This research was funded by an NSERC Discovery grant, Ontario Ministry of Innovation Early Researcher Award, and the Ontario Ministry of Environment Best in Science, York University Research Chair grants awarded to S.S. York University Faculty of Graduate Studies salary also supported O.M. We thank Gavin Simpson and two anonymous reviewers whose comments substantially improved the manuscript.
Author information
Authors and Affiliations
Contributions
D.G., S.S. and A.F. conceived the idea for generating a database. L.M., T.S., A.S., C.E., O.M. and A.F. collected the data. K.B., O.M., A.F. and D.G. made figures and analyzed the data. A.F. and O.M. wrote the initial draft. All authors participated in verifying the data and revising the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
The Creative Commons Public Domain Dedication waiver http://creativecommons.org/publicdomain/zero/1.0/ applies to the metadata files associated with this article.
About this article
Cite this article
Filazzola, A., Mahdiyan, O., Shuvo, A. et al. A database of chlorophyll and water chemistry in freshwater lakes. Sci Data 7, 310 (2020). https://doi.org/10.1038/s41597-020-00648-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-020-00648-2
This article is cited by
-
Consistent stoichiometric long-term relationships between nutrients and chlorophyll-a across shallow lakes
Nature Communications (2024)
-
High-resolution freshwater dissolved calcium and pH data layers for Canada and the United States
Scientific Data (2024)
-
Distribution of Antibiotic Resistance Genes and Their Associations with Bacterial Communities and Water Quality in Freshwater Lakes
Water, Air, & Soil Pollution (2023)
-
Indicators of the effects of climate change on freshwater ecosystems
Climatic Change (2023)
-
Imbalance of global nutrient cycles exacerbated by the greater retention of phosphorus over nitrogen in lakes
Nature Geoscience (2022)