Abstract
We present Chia, a novel, large annotated corpus of patient eligibility criteria extracted from 1,000 interventional, Phase IV clinical trials registered in ClinicalTrials.gov. This dataset includes 12,409 annotated eligibility criteria, represented by 41,487 distinctive entities of 15 entity types and 25,017 relationships of 12 relationship types. Each criterion is represented as a directed acyclic graph, which can be easily transformed into Boolean logic to form a database query. Chia can serve as a shared benchmark to develop and test future machine learning, rule-based, or hybrid methods for information extraction from free-text clinical trial eligibility criteria.
Measurement(s) | Clinical Trial Eligibility Criteria • Analytical Procedure Accuracy |
Technology Type(s) | digital curation • computational modeling technique |
Sample Characteristic - Organism | Homo sapiens |
Machine-accessible metadata file describing the reported data: https://doi.org/10.6084/m9.figshare.12765602
Similar content being viewed by others
Background & Summary
Clinical trial eligibility criteria specify rules for screening clinical trial participants and play a central role in clinical research in that they are interpreted, implemented, and adapted by multiple stakeholders at various phases in the clinical research life cycle1. After being defined by investigators, eligibility criteria are used and interpreted by clinical research coordinators for screening and recruitment. Then, they are used by query analysts and research volunteers for patient screening. Later, they are summarized in meta-analyses for developing clinical practice guidelines and, eventually, interpreted by physicians to screen patients for evidence-based care. Hence, eligibility criteria affect recruitment, results dissemination, and evidence synthesis.
Despite their importance, recent studies highlight the often negative impact these criteria have on the generalizability of a given trial’s findings in the real world2,3. When eligibility criteria lack population representativeness, the enrolled participants cannot unbiasedly represent those who will be treated according to the results from that study4. Given that eligibility criteria are written in free text, it is laborious to answer this representativeness question at scale5. A related challenge is to assess the comparability of trial populations, especially for multi-site studies: e.g.,, given two clinical trials investigating the same scientific question, can we tell if they are studying comparable cohorts? The manual labor required from domain experts for such appraisal is prohibitive. Another challenge is patient recruitment, or finding eligible patients for a clinical trial, which remains the leading cause of early trial termination6,7. Unsuccessful recruitment wastes financial investment and research opportunities, on top of missed opportunities, inconvenience, or frustration of patients when the clinical trial is terminated early or cancelled.
Computable representations of eligibility criteria promise to overcome the above challenges and to improve study feasibility and recruitment success8. The Biomedical Informatics research community has produced various knowledge representations for clinical trial eligibility criteria9, though nearly all of them predate the current state-of-the-art in machine learning, and some even predate contemporary electronic health records9. Early efforts to create annotated datasets in eligibility criteria have used a variety of methods including ad-hoc annotation10, manual annotation of standardized biomedical concepts11, as well as leveraging biomedical knowledge resources such as UMLS for automatic semantic pattern extraction12. The annotations in these datasets do not capture sufficient information to form the logical statements of a database query, and few annotated datasets are publicly available. Ross et al. published a dataset with 1,000 eligibility criteria and analyzed their semantic complexity, but the data were not amenable for machine learning13. 79 eligibility criteria were annotated by Weng et al. with semantic tags and relations, but these are too few to serve as a sufficiently large training resource12. The most robustly annotated and the only publicly available corpus to date was produced by Kang et al.14, who annotated eligibility criteria from 230 clinical trials, though all on Alzheimer’s Disease. Hence the corpus lacks generalizability to other diseases. These and other works have focused on bridging the gap between eligibility criteria and logical queries (Table 1), but the percentage of criteria that could be fully represented using these annotation models and used in database queries (here referred to as criteria coverage) is variable, ranging from 18% to 87%5,14,15,16,17,18.
A shared, sufficiently large dataset is much needed to boost machine learning natural language processing of eligibility criteria text. In this study we present Chia, a large annotated corpus of clinical research eligibility criteria from 1,000 diverse clinical studies. The annotations specify (a) the boundaries and semantic categories of named entities and (b) the Boolean operators needed to form the database query logic. As the first public large annotated corpus for clinical trial eligibility criteria, Chia can serve as a shared benchmark to develop and test future machine learning, rule-based, or hybrid methods for information extraction from free-text clinical trial eligibility criteria.
Methods
Chia’s Annotation Model (CAM)
The annotation model was developed by two annotators (FK and LHF), both with medical training, and one machine learning researcher (CY), following an iterative process. The entity categories are aligned with the domain names defined by the Observational Health Data Sciences and Informatics (OHDSI) OMOP Common Data Model (CDM), which is widely used in the medical research community for health data standardization19. Our annotation model is described in full in the Appendix. A brief description is provided below with a focus on its three main components: Entities, Relationships, and the resulting Annotation Graph.
Entities
Entities are concepts (e.g., hypertension, platelet count) and fall into one of the three types: Domain, Field, and Construct. Domain entities are the essential components of eligibility criteria, while Field entities and Construct entities are optional, depending on the semantic categories of Domain entities. Domain entities represent eight domains, i.e., observation, condition, person, device, drug, visit, procedure, and measurement. A Field entity represents a property of an applicable domain entity. For example, rule “Hemoglobin <8 g/dL” includes a Domain entity “Hemoglobin” and a Field entity “ <8 g/dL.” A Construct entity defines modifiers such as negation and repetition. For example, the Negation entity inverts the Boolean logic of the Domain entity being modified: no history of heart disease.
Relationships
Relationships express general Boolean algebra operators (AND or OR) between pairs of entities, as well as type-specific relationships (e.g.,, has_value, has_temporal etc.). This dataset represents 12 relationships: AND, OR, SUBSUMES, HAS_NEGATION (target argument is negation), HAS_MULTIPLIER (target argument is multiplier), HAS_QUALIFIER (target argument is qualifier), HAS_VALUE (target argument is value), HAS_TEMPORAL (target argument is temporal), HAS_INDEX (target argument is reference_point), HAS_MOOD (target argument is mood), HAS_CONTEXT (target argument is observation and not included in above relationships), and HAS_SCOPE (target argument is scope). The Boolean operator NOT, as mentioned in the previous section, is an entity instead of a relationship. When considering general Boolean relationships, both entities are independent, and the truth value of each term (i.e., is an entity “true”?) are resolved before applying Boolean logic to the relationship. For example, in “Patients with diabetes or hypertension,” “diabetes” and “hypertension” are independent Domain entities linked by an OR relationship between them.
Annotation graph
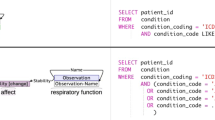
The entities and relationships in each trial’s eligibility criteria can be represented as two Annotation Graphs (AGs) (one for inclusion criteria and one for exclusion criteria) to computationally represent the query logic, that is, how all entities and relationships should be used to construct an executable query. The entities form the nodes and the relationships form the edges in the graph, and each entity evaluates to true if matching data can be found to satisfy it. Parsing of a graph runs from root to leaf nodes, forming a single Boolean expression. If that expression evaluates to true when fed the data from one patient, it means such patient is eligible (if inclusion criteria) or is ineligible (if exclusion criteria). An example annotation graph can be seen in Fig. 1.
Scope objects
As eligibility criteria are used to identify patients eligible for a given trial, complex logic is often employed to ensure clinical judgments can be made with a high degree of accuracy. In some criteria, this complex logic can be considered ‘nested’ as a single entity is explained in greater detail and is provided with additional parameters whereby the entity could evaluate to true. An example is “patient has hypertension (systolic blood pressure >135 mmHg or diastolic blood pressure >85 mmHg).” Here, the central entity is hypertension, which can be evaluated using the specified systolic and diastolic pressure ranges. We can treat both blood pressure measurements connected by an OR relationship as a single logical statement by labeling it a Scope object. Thus, this criterion can be rewritten to “patient has hypertension, or [systolic blood pressure > 135 mmHg or diastolic blood pressure > 85 mmHg].” The hypertension entity is thus linked to this Scope object whereby satisfying either item results in the statement resolving to a single value of True.
The annotation processes
Sampling of trials and loading into the annotation tool
We searched ClinicalTrials.gov on August 2, 2018 for actively recruiting, interventional (clinical trial), phase 4 studies, and obtained 2,913 trials, from which a random sample of 1,000 clinical trials was drawn. We focused on current studies as opposed to historic ones, assuming reporting quality is generally better in more current trials20, and prioritized phase 4 since they are more likely to be replicated via pure observational data analyses21. From each trial, a script downloaded and extracted every eligibility criterion (roughly defined as one line of free text), exported plain text files, and loaded them into the brat annotation tool (http://brat.nlplab.org/). That script is in R language and is available at https://github.com/WengLab-InformaticsResearch/CHIA.
Annotation of the eligibility criteria from the selected trials
The creation of Chia was performed by medical professionals (FK and LHF). Each annotator received a separate set of criteria loaded in brat22 and hand-created the entities and relationships as expressed above. In case of doubt for some concept, the annotator searched terminology at http://athena.ohdsi.org, which provides searching of concepts in the OMOP CDM. For the first 200 trials, both annotators regularly discussed adaptations to the annotation model based on their experience and re-annotated criteria as needed according to the changes being made. Once a satisfactory model was attained based on the consensus of both annotators, modifications were suspended and the task of annotation proceeded until the completion of the 1,000 trials. The final Chia dataset contains the summed and collectively revised work of the two annotators.
Post-processing the annotations
Minor post-production was performed to transform the ANN files produced by brat into a single long table in CSV format containing the entire dataset. That table also contains a number of variables that can be programmatically inferred from the annotations, e.g.,, which entities are roots in their annotation graphs. Additional post-processing was performed to generate two distinct datasets: one titled With Scopes and the other Without Scopes differing only in their utilization of Scope entity within the annotation model. Greater discussion of the reasoning behind the two distinct datasets is included in the Appendix, and all code used to generate these two models is available at https://github.com/WengLab-InformaticsResearch/CHIA.
To identify the target diseases of the 1,000 annotated trials, additional dataset enrichment was accomplished by leveraging the Aggregate Analysis of ClinicaTrials.gov (AACT) database23. This publicly available relational database contains all information about every study registered in ClinicalTrials.gov and is provided by the Clinical Trials Transformation Initiative (CTTI). The list of 1,000 unique NCT IDs included in our dataset was extracted and matched with their corresponding target conditions using the conditions table in the AACT database.
Parsing the annotation graph
A distinguishing feature of our dataset is its capacity to support the parsing of the entities and their relationships into a Boolean expression containing the logic of the database query that replicates the eligibility criteria of each clinical trial. A sample annotation alongside its associated annotation graph and pseudo-query is provided in Fig. 1.
Data Records
The free-text of selected eligibility criteria, brat configuration files, and the annotated data files are all available on figshare at https://doi.org/10.6084/m9.figshare.1185581724. There are two folders of annotation files titled With Scope and Without Scope, describing the inclusion or exclusion of Scope entities (additional information in Appendix).
Free-text (.txt) Files
Extracted free-text eligibility criteria from the 1,000 selected trials. Each text file adheres to the following naming format: [NCT Number][Inclusion/Exclusion Status].txt. Each row contains a single eligibility criterion.
Annotation (.ann) Files
Non-post-processed annotations in brat’s native ANN output format. Each annotation file adheres to the following naming format: [NCT Number]_ [Inclusion/ExclusionStatus].ann. As per the ANN format, each line corresponds to a single entity or relationship, except for OR relationships as explained below. For entities, the fields are as follows: item ID (e.g., T1), entity type (e.g., Condition), string start index (e.g., 28), string end index (e.g., 55), text (e.g., ‘metastatic carcinoid tumors’). For relationships, the fields are as follows: item ID (e.g., R1), relationship type (e.g., has_value), root argument (e.g., T3), target argument (e.g., T2). For OR relationships, the second field lists the relationship type and subsequent columns list all items connected by this OR relationship. In order to visualize the annotations, one needs brat. Simply open, in brat, Chia’s entire unzipped folder. Instructions for downloading and installing brat are available at http://brat.nlplab.org/.
Configuration (.conf) Files
These are the brat configuration files used to produce annotations following the definitions of CAM. They are used to load the.ann files into brat for viewing or editing them.
Technical Validation
Inter-annotator agreement
To evaluate the inter-annotator agreement of CHIA, we randomly selected 50 trials out of the 1,000 trials, constituting 5% of the whole dataset with 604 inclusion criteria and 1,034 exclusion criteria. The same 50 trials were provided to the two annotators (FK and AB) to annotate independently using the Brat annotation tool. To facilitate our agreement evaluation, the obtained raw annotations were converted into two formats: the CONLL-2003 BIO format25 and the token-level format with annotated label on each token. For example, in the CONLL-2003 BIO format, Diabetes mellitus is annotated as “diabetes B-Condition mellitus I-Condition”. In the token-level format, Diabetes mellitus is annotated as “diabetes Condition mellitus Condition”. An agreement was reached if both annotators annotated the whole span of Diabetes mellitus as Condition. These two formats enable us to evaluate both phrase-level agreement and token-level agreement rates. We calculated the Cohen’s kappa scores and Precision, Recall, and F1 as the measure of inter-annotator agreement. At the phrase-level, the Cohen’s kappa score is 0.8043 with Precision 81.39%, Recall 80.30% and F1 80.84. At the token-level, the Cohen’s kappa score is 0.8489 with Precision 81.67%, Recall 86.68% and F1 84.10.
Dataset exploration and validation
Descriptive statistics were generated to report counts of entities, relationships, and combinations of both. A coverage statistic was used to compare this dataset with previous efforts. In an effort to assess the accuracy and immediate utility of individual annotations, the raw unprocessed annotated entities were mapped to standardized medical concepts. To do this we utilized an open-source concept mapping tool called Usagi (https://www.ohdsi.org/web/wiki/doku.php?id=documentation:software:usagi)26 to map free-text strings to medical concepts in the OMOP CDM and to provide a ‘mapping accuracy score’ based on string similarity that is automatically generated by the Usagi tool. All Domain entities were converted to lowercase and then input into Usagi v1.2.7 with the following restrictions: (1) ‘Filter standard concepts’ was unselected to allow for mapping to Standard and Classification Concepts, (2) ‘Filter by Domain’ was selected and the selected options included only the labeled Domain (e.g., Condition).
Descriptive statistics
Chia contains 65,886 annotations for 12,409 inclusion and exclusion eligibility criteria from 1,000 trials targeting 1,130 unique diseases, with the most common one being Coronary Artery Disease (24 trials). 196 of the trials included healthy volunteers. 1,606 of the annotated eligibility criteria were determined as not being useful for patient-focused database queries for reasons highlighted in Table 2. Of those, 462 contained multiple entity annotations (e.g.,, “1. Signed informed consent” was annotated with parsing_error, non-query-able, and informed_consent) so the sum of annotations in Table 2 is greater than 1,606. 10,768 eligibility criteria had evaluable annotations, accounting for 86.8% of all the eligibility criteria (Table 1). When comparing it to the dataset prepared by Kang et al. in 201714 (called EliIE), Chia is larger in size in terms of number of annotations, number of entity and relationship types, and criteria coverage. A top-level comparison of these datasets is shown in Table 3.
Of the 41,699 entity annotations, 63.5% fall into the Domain category, 18.4% in the Field category, 17.5% in the Construct category and 0.4% were concepts unable to be annotated or deemed ‘unqueryable’ – additional information on these categories is included in the Appendix. Condition is the most common entity and OR the most common relationship. The most common relationship triplet is Measurement-has_value-Value; the second most common is Condition-has_qualifier-Qualifier. The most common relationship types are listed in Table 4 and the most common relationship triplets are listed in Table 5. OR relationships were not included in calculating the most common triplets as they do not always follow the root-relationship-target structure. 29.9% of OR relationships linked more than 2 entities and maximally linked 25 entities. All type-specific relationships contained the respective target entity type (e.g., has_value to Value entity) except for has_temporal (86.7% Temporal, 13.3% Observation) and has_mood (97.7% Mood, 2.3% Observation), though these different target types reflect the flexibility of the Observation Domain. For example, “history of” is considered an Observation despite its role in the has_temporal relationship. Concept mappings to the OMOP CDM with a score greater than or equal to 0.7 were considered strong matches. In total, 65.9% of raw annotated entities within the 8 Domain entity categories were considered strong mappings to the OMOP CDM. The percentage of strong mappings in each Domain category is shown in Table 6. Finally, the 15 most frequent tokens within the most common entity types are listed in Table 7 (fuzzy string-matching was used to generate token-level information).
The Chia dataset contains a total of 4,161 annotated Scope objects with 1,009 having an incoming subsumes or multi relationship, serving a body of original and useful knowledge for electronic phenotyping (discussed further in Use Case 2 below). A few examples of Scope objects are presented in Table 8. The average number of entities contained within these Scope objects is 3.51 with a maximum of 82. Additionally, 2,318 subsumes or multi relationships are available throughout the dataset with that number increasing to 2,521 in the dataset without Scope objects (post-Scope decomposition).
Usage Notes
Use case analyses
To demonstrate the utility of Chia, we propose two motivating use cases for this annotated corpus that can be explored in future research efforts.
Use Case 1: Machine learning model training for information extraction from eligibility criteria
Machine learning technologies can assist in parsing eligibility criteria. In previous research, Conditional Random Fields (CRFs)27, Convolutional Neural Networks (CNNs)28, Support Vector Machines (SVMs)14, hierarchical clustering29, distant supervision30, and other machine learning approaches have been used to extract entities and relationships from the free-text eligibility criteria to obtain structured representations. These extraction methods typically result in satisfactory accuracy but relatively low recall, such as the 94% accuracy and 18% recall described by Milian et al. in 201517. There are a few possible explanations for this low recall. The first is that the training corpus lacks diversity in the types of eligibility criteria, as described in more detail in the Background section of this article. The second explanation is that most of the criteria (~85%) are very complex with various semantic patterns, including 35% of criteria containing more than one type of semantic pattern13,31. Finally, incomplete sentence structure and word ambiguity can also lead to extraction failures as described by Yuan et al. in 201932.
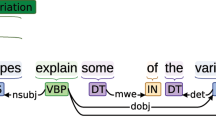
A larger dataset of eligibility criteria with samples from a broad range of clinical trials is necessary to train a more accurate and robust extraction model. With 41,699 entities and 25,017 relationships annotated, Chia provides ample training data for machine learning research for identifying not only the boundaries and classes of named entities within medical text, but also the dependencies between these concepts. Further, when comparing to previous annotation efforts outlined in the Background section, the richness of the Chia model becomes clear as highlighted in Fig. 2. This direct criterion-to-criterion comparison allows for better appreciation of the increased level of connectivity between concepts (section A), simplicity in structure and format (section B), and improved granularity (section C) provided by Chia.
Chia’s non-flat annotation scheme is also noteworthy. Most corpuses for Named Entity Recognition (NER) training have adopted the flat annotation, disallowing discontinuous, nested, or overlapping entities33, whereas Chia represents them and supports the use case Electronic Phenotyping described below. Discontinuous and overlapping entities are required to capture coordination ellipsis, such as “presence of pulmonary, hepatic or hematological cancer.”, which is one type of ellipsis used in coordinating structure to achieve textual concision by omitting repeated words34. Coordination ellipses are more frequently used in medical language than in the general domain. More granular results are required for downstream tasks such as free-text based phenotyping. Existing annotated corpus containing overlapping entities are derived from biomedical literature, including GENIA and NCBI Disease corpora35. GENIA corpus is focused primarily on biological entities such as DNA, RNA, and protein36, and NCBI Disease is focused on disease mentions. To the best of our knowledge, Chia is the first clinical corpus of considerable size that is annotated in a non-flat fashion and supports annotation nesting and joining.
Use Case 2: Electronic phenotyping
Electronic phenotyping plays an essential role in disease knowledge discovery, application, and clinical research2,37. Electronic phenotyping supports cross-sectional and epidemiological studies, association studies, clinical risk factor analyses, and cohort identification2. In some cases, the phenotype definition is fairly simple (e.g., diagnosis of rheumatoid arthritis), but it can become more nuanced and complex (e.g., moderate or severe COPD exacerbation [requiring corticosteroids or increased dosage of corticosteroids and/or antibiotics or hospitalization]). Manual knowledge engineering to establish the linkage between the concepts in a phenotype is neither scalable nor efficient. Hierarchical relationships between annotated entities are explicitly defined in Chia via Scope objects, subsumes relationships and multi relationships, which provide reusable phenotyping knowledge. These annotations often indicate one concept (or group of concepts) that can be substituted for another because their meanings are inherently interconnected – oftentimes because the latter are clarifications or specifications of the former. For example, Systolic Blood Pressure > 130 and Diastolic Blood Pressure >85 can be used to define the condition Hypertension. There are 2,197 cases of subsumes in Chia, and Table 9 gives a few examples. Further, as eligibility criteria themselves serve to define a patient cohort, they can be considered to be small electronic phenotypes.
Code availability
All code used to generate and process the datasets described above is freely available at https://github.com/WengLab-InformaticsResearch/CHIA in the file titled chia.R. It was written in R version 3.3.3.
References
Weng, C. Optimizing Clinical Research Participant Selection with Informatics. Trends in pharmacological sciences 36, 706–709, https://doi.org/10.1016/j.tips.2015.08.007 (2015).
Banda, J. M., Seneviratne, M., Hernandez-Boussard, T. & Shah, N. H. Advances in Electronic Phenotyping: From Rule-Based Definitions to Machine Learning Models. Annual review of biomedical data science 1, 53–68, https://doi.org/10.1146/annurev-biodatasci-080917-013315 (2018).
Sen, A. et al. Correlating eligibility criteria generalizability and adverse events using Big Data for patients and clinical trials. Ann N Y Acad Sci 1387, 34–43, https://doi.org/10.1111/nyas.13195 (2017).
Murthy, V. H., Krumholz, H. M. & Gross, C. P. Participation in cancer clinical trials: race-, sex-, and age-based disparities. Jama 291, 2720–2726, https://doi.org/10.1001/jama.291.22.2720 (2004).
Chondrogiannis, E. et al. A novel semantic representation for eligibility criteria in clinical trials. Journal of biomedical informatics 69, 10–23, https://doi.org/10.1016/j.jbi.2017.03.013 (2017).
Williams, R. J., Tse, T., DiPiazza, K. & Zarin, D. A. Terminated Trials in the ClinicalTrials.gov Results Database: Evaluation of Availability of Primary Outcome Data and Reasons for Termination. PloS one 10, e0127242, https://doi.org/10.1371/journal.pone.0127242 (2015).
Richesson, R. L. et al. Electronic health records based phenotyping in next-generation clinical trials: a perspective from the NIH Health Care Systems Collaboratory. J Am Med Inform Assoc 20, e226–e231, https://doi.org/10.1136/amiajnl-2013-001926 (2013).
Weng, C. Optimizing Clinical Research Participant Selection with Informatics. Trends Pharmacol Sci 36, 706–709, https://doi.org/10.1016/j.tips.2015.08.007 (2015).
Weng, C., Tu, S. W., Sim, I. & Richesson, R. Formal representation of eligibility criteria: a literature review. Journal of biomedical informatics 43, 451–467, https://doi.org/10.1016/j.jbi.2009.12.004 (2010).
Patel, P., Davey, D., Panchal, V. & Pathak, P. Annotation of a Large Clinical Entity Corpus. (2018).
Mohan, S. & Li, D. MedMentions: A Large Biomedical Corpus Annotated with UMLS Concepts. (2019).
Weng, C. et al. EliXR: an approach to eligibility criteria extraction and representation. Journal of the American Medical Informatics Association 18, i116–i124, https://doi.org/10.1136/amiajnl-2011-000321 (2011).
Ross, J., Tu, S., Carini, S. & Sim, I. Analysis of eligibility criteria complexity in clinical trials. Summit Transl Bioinform, 46–50 (2010).
Kang, T. et al. EliIE: An open-source information extraction system for clinical trial eligibility criteria. J Am Med Inform Assoc 24, 1062–1071, https://doi.org/10.1093/jamia/ocx019 (2017).
Tu, S. W. et al. A practical method for transforming free-text eligibility criteria into computable criteria. Journal of biomedical informatics 44, 239–250, https://doi.org/10.1016/j.jbi.2010.09.007 (2011).
Zhang, H. et al. Computable Eligibility Criteria through Ontology-driven Data Access: A Case Study of Hepatitis C Virus Trials. AMIA Annu Symp Proc, 1601–1610 (2018).
Milian, K. et al. Enhancing reuse of structured eligibility criteria and supporting their relaxation. Journal of biomedical informatics 56, 205–219, https://doi.org/10.1016/j.jbi.2015.05.005 (2015).
Lonsdale, D., Tustison, C., Parker, C. & Embley, D. Formulating Queries for Assessing Clinical Trial Eligibility. (2006).
Reich, C., Ryan, P. B., Belenkaya, R., Natarajan, K. & Blacketer, C. OHDSI Common Data Model v6.0 Specifications, https://github.com/OHDSI/CommonDataModel/wiki (2019).
Zarin, D. A., Fain, K. M., Dobbins, H. D., Tse, T. & Williams, R. J. 10-Year Update on Study Results Submitted to ClinicalTrials.gov. New England Journal of Medicine 381, 1966–1974, https://doi.org/10.1056/NEJMsr1907644 (2019).
Suvarna, V. Phase IV of Drug Development. Perspect Clin Res 1, 57–60 (2010).
Stenetorp, P. et al. brat: a Web-based Tool for NLP-Assisted Text Annotation. Proceedings of the Demonstrations at the 13th Conference of the European Chapter of the Association for Computational Linguistics, 102–107 (2012).
Clinical Trials Transformation Initiative. Aggregate Analysis of ClinicalTrials.gov, https://aact.ctti-clinicaltrials.org/ (2016).
Kury, F. S. P. et al. Chia Annotated Datasets. figshare https://doi.org/10.6084/m9.figshare.11855817.v2 (2020).
Sang, E. F. & De Meulder, F. Introduction to the CoNLL-2003 shared task: Language-independent named entity recognition. arXiv preprint cs/0306050 (2003).
Observational Health Data Sciences and Informatics. Usagi, https://www.ohdsi.org/web/wiki/doku.php?id=documentation:software:usagi (2018).
Luo, Z., Johnson, S. B., Lai, A. M. & Weng, C. Extracting temporal constraints from clinical research eligibility criteria using conditional random fields. AMIA Annu Symp Proc, 843–852 (2011).
Chuan, C.-H. Classifying Eligibility Criteria in Clinical Trials Using Active Deep Learning. (2018).
Luo, Z., Johnson, S. B. & Weng, C. Semi-Automatically Inducing Semantic Classes of Clinical Research Eligibility Criteria Using UMLS and Hierarchical Clustering. AMIA Annu Symp Proc, 487–491 (2010).
Sun, Y. & Loparo, K. In 2019 IEEE 43rd Annual Computer Software and Applications Conference (COMPSAC). 954–955.
Sun, Y. & Loparo, K. Knowledge-guided Text Structuring in Clinical Trials. (2019).
Yuan, C. et al. Criteria2Query: a natural language interface to clinical databases for cohort definition. J Am Med Inform Assoc 26, 294–305, https://doi.org/10.1093/jamia/ocy178 (2019).
Alex, B., Haddow, B. & Grover, C. Recognising nested named entities in biomedical text. (Association for Computational Linguistics, 2007).
Yuan, C. et al. A Graph-Based Method for Reconstructing Entities from Coordination Ellipsis in Medical Text. Journal of the American Medical Informatics Association (2020).
Doğan, R. I., Leaman, R. & Lu, Z. NCBI disease corpus: a resource for disease name recognition and concept normalization. Journal of biomedical informatics 47, 1–10, https://doi.org/10.1016/j.jbi.2013.12.006 (2014).
Kim, J. D., Ohta, T., Tateisi, Y. & Tsujii, J. GENIA corpus–semantically annotated corpus for bio-textmining. Bioinformatics 19(Suppl 1), i180–182, https://doi.org/10.1093/bioinformatics/btg1023 (2003).
Banda, J. M., Halpern, Y., Sontag, D. & Shah, N. H. Electronic phenotyping with APHRODITE and the Observational Health Sciences and Informatics (OHDSI) data network. AMIA Jt Summits Transl Sci Proc, 48–57 (2017).
Acknowledgements
This project was supported by The National Library of Medicine grant R01LM009886-10 (“Bridging the semantic gap between research eligibility criteria and clinical data”) and the National Center for Advancing Translational Science (NCATS) grant UL1TR001873.
Author information
Authors and Affiliations
Contributions
Fabrício Kury – Primary dataset annotator, drafted original manuscript. Li-heng Fu – Secondary dataset annotator, aided in revision of manuscript. Alex Butler – Assisted with dataset annotation, aided in revision of manuscript. Chi Yuan – Machine learning developer, produced results presented in Technical Validation section. Yingcheng Sun – aided in revision of manuscript. Hao Liu – Programmer, developed code used for Scope decomposition. Ida Sim – Helped develop and refine the details of CAM, aided in reviewing the results. Simona Carini – Helped develop and refine the details of CAM, aided in reviewing the results. Chunhua Weng – Research supervisor, principal investigator, conceptualization and research design of the study, guarantor for the annotated datasets, review, editing, and revision of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
The Creative Commons Public Domain Dedication waiver http://creativecommons.org/publicdomain/zero/1.0/ applies to the metadata files associated with this article.
About this article
Cite this article
Kury, F., Butler, A., Yuan, C. et al. Chia, a large annotated corpus of clinical trial eligibility criteria. Sci Data 7, 281 (2020). https://doi.org/10.1038/s41597-020-00620-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-020-00620-0
This article is cited by
-
Improving clinical trial design using interpretable machine learning based prediction of early trial termination
Scientific Reports (2023)
-
A review of research on eligibility criteria for clinical trials
Clinical and Experimental Medicine (2023)
-
A comparative study of pre-trained language models for named entity recognition in clinical trial eligibility criteria from multiple corpora
BMC Medical Informatics and Decision Making (2022)
-
ELaPro, a LOINC-mapped core dataset for top laboratory procedures of eligibility screening for clinical trials
BMC Medical Research Methodology (2022)
-
The Leaf Clinical Trials Corpus: a new resource for query generation from clinical trial eligibility criteria
Scientific Data (2022)