Abstract
The Assay for Transposase-Accessible Chromatin using sequencing (ATAC-seq) is a fundamental epigenomics approach and has been widely used in profiling the chromatin accessibility dynamics in multiple species. A comprehensive reference of ATAC-seq datasets for mammalian tissues is important for the understanding of regulatory specificity and developmental abnormality caused by genetic or environmental alterations. Here, we report an adult mouse ATAC-seq atlas by producing a total of 66 ATAC-seq profiles from 20 primary tissues of both male and female mice. The ATAC-seq read enrichment, fragment size distribution, and reproducibility between replicates demonstrated the high quality of the full dataset. We identified a total of 296,574 accessible elements, of which 26,916 showed tissue-specific accessibility. Further, we identified key transcription factors specific to distinct tissues and found that the enrichment of each motif reflects the developmental similarities across tissues. In summary, our study provides an important resource on the mouse epigenome and will be of great importance to various scientific disciplines such as development, cell reprogramming, and genetic disease.
Design Type(s) | epigenetic modification identification objective • organism part comparison design |
Measurement Type(s) | assay for transposase-accessible chromatin using sequencing |
Technology Type(s) | next generation DNA sequencing |
Factor Type(s) | technical replicate • biological replicate • animal body part |
Sample Characteristic(s) | Mus musculus • adrenal gland • brain • cerebellum • visceral abdominal adipose tissue • brown adipose tissue • adipose tissue • heart • kidney • liver • lung • ovary • pancreas • skeletal muscle tissue • spleen • thymus • uterus • bladder organ • large intestine • small intestine • stomach |
Machine-accessible metadata file describing the reported data (ISA-Tab format)
Similar content being viewed by others
Background & Summary
Although most of the protein-coding genes in human and model animals such as mouse have been extensively annotated, vast regions of the genome are noncoding sequences (e.g., roughly 98% of the human genome) and still poorly understood1,2. During the last decade, the development of next-generation sequencing (NGS) based epigenomics techniques (e.g., ChIP-seq and DNase-seq) have significantly facilitated the identification of functional genomic regions3. For example, by comparing the histone modifications and transcription factor (TF) binding patterns throughout the mouse genome in a wide spectrum of tissues and cell types, Yue et al.4,5 have made significant progress towards a comprehensive catalog of potential functional elements in the mouse genome. So far, the international human epigenome consortium (IHEC), including ENCODE and the NIH Roadmap epigenomics projects, have profiled thousands of epigenomes including DNA methylation, genome-wide binding of TFs, histone modifications, and chromatin accessibility. This has resulted in the discovery of over 5 million cis-regulatory elements (CREs) in the human genome6,7,8. These data resources have created an important baseline for further study of diverse biological processes, such as development, cell reprogramming, and human disease9,10,11,12,13.
The accessibility of CREs, which is important for switching on and off gene expression14, is strongly associated with transcriptional activity. To date, detection of DNase I hypersensitive sites (DHSs) within chromatin by DNase-seq has been extensively used to map accessible genomic regions in diverse organisms including the laboratory mouse5. In 2013, Buenrostro et al.15 reported an alternative approach, termed ATAC-seq, for fast and sensitive profiling of chromatin accessibility by direct transposition of native chromatin within the nucleus. This method, in comparison to DNase-seq, requires a significantly lower input of cells (only 500–50,000) and a shorter period to process samples16. Moreover, ATAC-seq has been applied to single cells through various methods17,18,19,20, enabling the investigation of regulatory heterogeneity within complex tissues. As such, ATAC-seq has demonstrated great potential to be a leading method in assaying accessible chromatin genome-wide.
The sequence preference of both DNase I and Tn5 enzymes produced distinct but inevitable biases in DNase-seq and ATAC-seq21, making it impractical to directly compare datasets generated from the two methods. Therefore, although the DNase-seq atlas of adult mouse tissues has been published5, a baseline of chromatin accessible regions generated from ATAC-seq is still important for ATAC-seq based studies. Here, we applied Omni-ATAC-seq22, an approach that enables profiling of accessible chromatin from frozen tissues, to the generation of 66 chromatin accessibility datasets from 20 different tissues derived from both adult male and female C57BL/6J mice (Fig. 1a). Systematic analysis of the dataset identified a total of 296,574 accessible elements, of which 26,916 showed highly tissue-specific patterns. We further predicted TFs specific to distinct tissues and importantly, many of these have been validated by previous studies23,24,25,26,27. In this study, we provide a valuable resource, which can be used to elucidate transcriptional regulation and may further help understand diseases caused by regulatory dysfunction.
Methods
Sample collection
All relevant procedures involving animals were approved by the Institutional Review Board on Ethics Committee of BGI (Permit No. BGI-R085-1). C57BL/6J male and female mice were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd (Beijing, China). 8-week old mice were used for this study. Mice were housed under standard conditions of a specific pathogen-free, temperature-controlled environment with a 12-h day/night cycle28. The mice were sacrificed by cervical dislocation. Whole organs were extracted and cut into 2–3 pieces, respectively (50–200 mg/piece). Each sample was then quickly frozen in liquid nitrogen and stored at −80 °C until nuclei extraction was performed. In this study, we used 20 different organs or tissues, including adrenal gland, bladder, brain (cerebrum and cerebellum), fat (abdominal, brown and mesenteric), heart, intestine (large and small), kidney, liver, lung, ovary, pancreas, skeletal muscle, spleen, stomach, thymus, and uterus (as listed in Table 1).
Library construction and sequencing
Tissues were homogenized in a 2 ml Dounce homogenizer (with a loose and then tight pestle) with 10–20 strokes in 2 ml of 1X homogenization buffer on ice22. 400 μl of this nuclei suspension was transferred to a round-bottom 2 ml Lo-Bind Eppendorf tube for density gradient centrifugation following the protocol by Corces et al.22. After centrifugation, the nuclei band (about 200 μl) was collected, stained with DAPI, and nuclei were counted. Approximately 20,000–100,000 nuclei were transferred into a fresh tube and diluted in 1 ml ATAC-RSB +0.1% Tween-20 (Sigma-Aldrich, Darmstadt, Germany). Nuclei were centrifuged and the supernatant was carefully aspirated. Nuclei were treated in 50 μl transposition reaction mixture containing 10 mM TAPS-NaOH (pH 8.5), 5 mM MgCl2, 10% DMF, 2.5 μl of in-house Tn5 transposase (0.8 U/μl), 0.01% digitonin (Sigma-Aldrich, Darmstadt, Germany), 0.1% Tween-20, 31.5 μl of PBS, and 5 μl of nuclease-free water for 30 mins at 37 °C. Afterward, the DNA was purified with MinElute Purification Kit (Qiagen, Venlo, Netherlands) and amplified with primers containing barcodes, as previously described22,29.
All libraries were adapted for sequencing on the BGISEQ-500 platform30. In brief, the DNA concentration was determined by Qubit 3.0 (ThermoFisher, Waltham, MA). Pooled samples were used to make single-strand DNA circles (ssDNA circles). DNA nanoballs (DNBs) were generated from the ssDNA circles by rolling circle replication as previously described30. The DNBs were loaded onto patterned nano-arrays and sequenced on the BGISEQ-500 sequencing platform with paired end 50 base reads.
Preprocessing of the ATAC-seq datasets
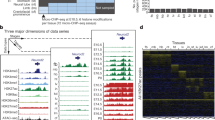
The ATAC-seq data were processed (trimmed, aligned, filtered, and quality controlled) using the ATAC-seq pipeline from the Kundaje lab31,32 (Table 2). The model-based analysis of ChIP-seq (MACS2)33 version 2.1.2 was used to identify the peak regions with options -B, -q 0.01–nomodel, -f BAM, -g mm. The Irreproducible Discovery Rate (IDR) method34 was used to identify reproducible peaks between two technical replicates (Fig. 1b). Only peaks reproducible between the two technical replicates were retained for downstream analyses. Peaks for all tissues were then merged together into a standard peak list. The number of raw reads mapped to each standard peak were counted using the intersect function of BedTools35 version 2.26.0. The raw count matrix32 was normalized by Reads Per Million mapped reads (RPM). Pearson correlation coefficients between technical or biological replicates across tissues were calculated based on the Log10 RPM matrix.
Identification of tissue-specific chromatin accessible regions
We used a strategy described previously based on the Shannon entropy to compute a tissue specificity index for each peak4,36,37. Specifically, for each peak, we defined its relative accessibility in a tissue type i as Ri = Ei/ΣE, where Ei is the RPM value for the peak in the tissue i, ΣE is the sum of RPM values in all tissues, and N is the total number of tissues. The entropy score for each peak across tissues can be defined as H = −1 * sum(Ri * log2Ri) (1 < i < N), where the value of H ranges between 0 to log2(N). An entropy score close to zero indicates the accessibility of this peak is highly tissue-specific, while an entropy score close to log2(N) indicates that this peak is ubiquitously accessible38. Based on the distribution of entropy scores, peaks with score less than 3.5 were selected as tissue-restricted peaks.
We searched TF motifs in tissue-specific peaks using the findMotifsGenome.pl script of the HOMER39 version 4.9.1 software with default settings. We then generated a motif enrichment matrix32, where each row represents the P-value of a motif and each column represents a tissue. The 50 motifs with the top CV values and mean values greater than 20 were displayed.
Data Records
A complete list of the 66 tissue samples is given in Tables 1 and 2. All raw data have been submitted to the CNGB Nucleotide Sequence Archive40. The raw data have also been submitted to the NCBI Sequence Read Archive41. The ATAC-seq QC results and count matrixes have been submitted to the Figshare32.
Technical Validation
Data QC from the pipeline with IDR quality control
We evaluated our ATAC-seq dataset by a series of commonly used statistics, including the number of total read, mapping rate, the proportion of duplicate read, the number of mitochondrial read, and the number of final usable read (Table 2). For each replicate, we obtained an average of 78 million reads, which was previously shown to be enough for the detection of accessible regions15,31. In agreement with published ATAC-seq profiles15, the chromatin accessibility fragments show size periodicity corresponding to integer multiples of nucleosomes32 (Fig. 2b). The successful detection of accessible regions is also supported by the observation of strong enrichment of ATAC-seq reads around transcription start sites (TSSs)32 (Fig. 2a,d).
ATAC-seq data quality metrics. (a) The ATAC-seq signal enrichment around the transcription start sites (TSSs) for 4 representative samples (kidney or spleen of male or female mice). (b) The insert size distribution of ATAC-seq profiles for the same samples shown in 2a. (c) The irreproducible discovery rate (IDR) analyses of ATAC-seq peaks for the indicated samples. The scatter plots show points for every peak, with their location representing the rank in each replicate. (d) Genome browser views of ATAC-seq signal for the indicated housekeeping gene (Gapdh) and tissue-specific genes.
To evaluate the reproducibility of accessible element discovery between replicates, we identified accessible regions in both replicates by using the MACS233 algorithm. We then applied the IDR method34 to find peaks that were reproducible between replicates in each tissue type (Fig. 2c). We identified an average of 43,421 reproducible peaks (Table 2). For downstream analyses, we filtered out low-quality data where the TSS enrichment scores are less than 10.0 and the number of reproducible peaks are less than 10,000.
Reproducibility of biological samples and comparison with published studies
The Pearson correlation analysis was used to further examine the reproducibility of biological and technical replicates. Heatmap clustering of Pearson correlation coefficients from the comparison of 66 datasets revealed a strong correlation between replicates of the same tissue (Fig. 3b), but a lower correlation between profiles from distinct tissues. This result is also supported by t-distributed stochastic neighbor embedding (t-SNE) analysis with tissue-restricted peaks of all profiles (Fig. 3a,c). Interestingly, correlations between replicates from mice of the same gender are generally higher than those from the opposite gender. This can be seen in the cerebrum where the correlation coefficient between replicates of female mice is 0.99 (Fig. 3d), while the coefficient between male and female is slightly reduced (0.96). We also compared our data to ATAC-seq profiles of postnatal mouse (day 0) tissues downloaded from ENCODE project42,43. Importantly, we found that both heart and lung were comparable with each other (Fig. 3e). Taken together, these analyses strongly suggest that our ATAC-seq profiles can reliably detect accessible chromatin regions in the mouse genome and can be used as a basic reference ATAC-seq dataset for future studies.
Evaluation of reproducibility across the ATAC-seq datasets. (a) The distribution curve of peak entropy scores. (b) Heatmap clustering of correlation coefficients across all 66 tissue ATAC-seq profiles. (c) t-SNE plot of all 66 ATAC-seq profiles based on the 26,916 tissue-specific peaks. (d) Scatter plots showing the Pearson correlations between technical (left) and biological (right) replicates. (e) Scatter plots showing the Pearson correlations between ENCODE postnatal mouse (day 0) datasets and our ATAC-seq profiles.
Inferring tissue-specific transcription factors
In an effort to validate the tissue-specific TF motifs identified in our dataset, we compared them to results from previous studies. Log2 RPM of the tissue-restricted peaks was shown in the heatmap (Fig. 4a). For example, we observed high enrichment of the NeuroG2 motif in cerebellum and cerebrum (Fig. 4b), in agreement with the role of NeuroG2 in controlling the temporal switch from neurogenesis to gliogenesis and regulating laminar fate transitions23. In brown fat, we found the CCAAT-enhancer-binding proteins (CEBP) motif to be highly specific (Fig. 4b). This is supported by a previous study demonstrating that CEBP can cooperate with PRDM16 to induce brown fat determination and differentiation24. In addition, other well-known tissues-specific motifs such as the liver-specific HNF family TF motifs (Hnf1, HNF1b, and HNF4a)25 and heart or skeletal muscle specific Mef2 family motifs (Mef2a, Mef2b, Mef2c, Mef2d)26,27 were validated in our study. To further validate whether the overall motif enrichment in each tissue can reflect tissue specificity we performed hierarchical clustering of tissues using Euclidean distance (Fig. 4c). This provided a result similar to hierarchical clustering of various mouse tissues based on RNA-seq data44. In addition, examination of tissues from the gastrointestinal (GI) tract (i.e., large intestine, small intestine, and stomach) showed tight clustering (Fig. 4c), which is likely due to their common functions such as lipid metabolism and energy hemostasis45,46. Skeletal muscle and heart tissue are found in the same branch, suggesting that patterns of chromatin accessibility in the two tissues are highly influenced by shared TF motifs such as those from the Mef2 family45.
Identification of tissue-specific chromatin accessible elements and transcription factors. (a) Heatmap clustering showing the tissue-specific accessible elements. (b) Enrichment of the indicated TF motifs in each tissue. The size and color of each point represent the motif enrichment P-value (−log10 P-value). (c) The hierarchical clustering of transcription factor enrichment scores in each tissue. Euclidian distances are shown in the legend.
Usage Notes
The ATAC-seq data processing pipeline, including read mapping, peak calling, IDR analysis, and read counting were run on the Linux operating system. The optimized parameters are provided in the main text. All R source codes used for the downstream data analyses and visualization are provided in Supplementary File 1.
Code Availability
The R codes used for correlation analysis, identification of tissue-specific chromatin accessible regions, and tissue-specific TFs are available in the supplementary materials (Supplementary File 1). A repository list containing the chromatin accessibility raw count matrix and the motif enrichment matrix is available online32.
References
Consortium, I. H. G. S. Initial sequencing and analysis of the human genome. Nature 409, 860 (2001).
Venter, J. C. et al. The sequence of the human genome. Science 291, 1304–1351 (2001).
Rivera, C. M. & Ren, B. Mapping human epigenomes. Cell 155, 39–55 (2013).
Shen, Y. et al. A map of the cis-regulatory sequences in the mouse genome. Nature 488, 116 (2012).
Yue, F. et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature 515, 355–364 (2014).
Consortium, E. P. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57 (2012).
Kundaje, A. et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317 (2015).
Stunnenberg, H. G. et al. The International Human Epigenome Consortium: a blueprint for scientific collaboration and discovery. Cell 167, 1145–1149 (2016).
Xu, Q. & Xie, W. Epigenome in Early Mammalian Development: Inheritance, Reprogramming and Establishment. Trends Cell Biol 28.3, 237–253 (2017).
Eckersley-Maslin, M. A., Alda-Catalinas, C. & Reik, W. Dynamics of the epigenetic landscape during the maternal-to-zygotic transition. Nat. Rev. Mol. Cell Biol. 19, 436–450 (2018).
Apostolou, E. & Hochedlinger, K. Chromatin dynamics during cellular reprogramming. Nature 502, 462 (2013).
Takahashi, K. & Yamanaka, S. A decade of transcription factor-mediated reprogramming to pluripotency. Nat. Rev. Mol. Cell Biol. 17, 183 (2016).
Karczewski, K. J. & Snyder, M. P. Integrative omics for health and disease. Nat. Rev. Genet. 19, 299 (2018).
Li, B., Carey, M. & Workman, J. L. The role of chromatin during transcription. Cell 128, 707–719 (2007).
Buenrostro, J. D., Giresi, P. G., Zaba, L. C., Chang, H. Y. & Greenleaf, W. J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013).
Meyer, C. A. & Liu, X. S. Identifying and mitigating bias in next-generation sequencing methods for chromatin biology. Nat. Rev. Genet. 15, 709 (2014).
Buenrostro, J. D. et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523, 486 (2015).
Cusanovich, D. A. et al. Multiplex single-cell profiling of chromatin accessibility by combinatorial cellular indexing. Science 348, 910–914 (2015).
Mezger, A. et al. High-throughput chromatin accessibility profiling at single-cell resolution. Nat. Commun. 9, 3647 (2018).
Chen, X., Natarajan, K. N. & Teichmann, S. A. A rapid and robust method for single cell chromatin accessibility profiling. Nat. Commun. 9, 5345 (2018).
Karabacak Calviello, A., Hirsekorn, A., Wurmus, R., Yusuf, D. & Ohler, U. Reproducible inference of transcription factor footprints in ATAC-seq and DNase-seq datasets using protocol-specific bias modeling. Genome Biol. 20, 42 (2019).
Corces, M. R. et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods 14, 959–962 (2017).
Dennis, D. J. et al. Neurog2 and Ascl1 together regulate a postmitotic derepression circuit to govern laminar fate specification in the murine neocortex. Proc.Natl Acad. Sci. 114, 4934–4943 (2017).
Kajimura, S. et al. Initiation of myoblast to brown fat switch by a PRDM16–C/EBP-β transcriptional complex. Nature 460, 1154 (2009).
Xanthopoulos, K. G. et al. The different tissue transcription patterns of genes for HNF-1, C/EBP, HNF-3, and HNF-4, protein factors that govern liver-specific transcription. Proc. Natl Acad. Sci 88, 3807–3811 (1991).
Fickett, J. W. Quantitative discrimination of MEF2 sites. Mol. Cell. Biol. 16, 437–441 (1996).
Lu, J., McKinsey, T. A., Zhang, C.-L. & Olson, E. N. Regulation of skeletal myogenesis by association of the MEF2 transcription factor with class II histone deacetylases. Mol. Cell 6, 233–244 (2000).
Fischer, A. W., Cannon, B. & Nedergaard, J. Optimal housing temperatures for mice to mimic the thermal environment of humans: An experimental study. Mol. Metab 7, 161–170 (2018).
Shang, Z. et al. Single-cell RNA-seq reveals dynamic transcriptome profiling in human early neural differentiation. Gigascience 7, 1–19 (2018).
Huang, J. et al. A reference human genome dataset of the BGISEQ-500 sequencer. Gigascience 6, 1–9 (2017).
Koh, P. W. et al. An atlas of transcriptional, chromatin accessibility, and surface marker changes in human mesoderm development. Sci. Data 3, 160109 (2016).
Liu C.-Y. et al. An ATAC-seq atlas of chromatin accessibility in mouse tissues. figshare, https://doi.org/10.6084/m9.figshare.c.4436264.v1 (2019).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Li, Q., Brown, J. B., Huang, H. & Bickel, P. J. Measuring reproducibility of high-throughput experiments. Ann. Appl. Stat 5, 1752–1779 (2011).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Barrera, L. O. et al. Genome-wide mapping and analysis of active promoters in mouse embryonic stem cells and adult organs. Genome Res. 18, 46–59 (2008).
Schug, J. et al. Promoter features related to tissue specificity as measured by Shannon entropy. Genome Biol. 6, R33 (2005).
Xie, W. et al. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell 153, 1134–1148 (2013).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
CNGB Nucleotide Sequence Archive https://db.cngb.org/cnsa/project/CNP0000198/public/ (2018).
NCBI Sequence Read Archive http://identifiers.org/ncbi/insdc.sra:SRP167062 (2018).
ENCODE https://www.encodeproject.org/experiments/ENCSR451NAE/ (2017).
ENCODE https://www.encodeproject.org/experiments/ENCSR102NGD/ (2017).
Söllner, J. F. et al. An RNA-Seq atlas of gene expression in mouse and rat normal tissues. Sci. Data 4, 170185 (2017).
Zhou, Q. et al. A mouse tissue transcription factor atlas. Nat. Commun. 8, 15089 (2017).
Sonawane, A. R. et al. Understanding tissue-specific gene regulation. Cell Rep. 21, 1077–1088 (2017).
Acknowledgements
We thank all members of the Cell and Development Lab (BGI) for helpful comments and Jian Zhang and Jie Chen from Shenzhen Institutes of Advanced Technology for assistance with sample collection. This work was supported by the National Key R&D Program of China (No. 2016YFC1303902), the Shenzhen Peacock Plan (No. KQTD20150330171505310), and the Science, Technology and Innovation Commission of Shenzhen Municipality (No. JCYJ20160531194327655).
Author information
Authors and Affiliations
Contributions
C.L., L.L., M.W. and X.W. conceived the idea. C.L. and M.W. collected samples. M.W. and C.L. generated the data. J.X., J. Xia, J.L., T.F., M.C. and Y.Y. assisted with the experiments. X.W. analyzed the data with the assistance of C.L., X.D., P.Z. and L.W.. C.L. wrote the manuscript with the input of X.W. and M.W.. L.L. and B.A.P. supervised the study and revised the manuscript. X.Z., F.C., W.Z., A.C. and Z.S. provided helpful comments on this study. All authors reviewed and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
ISA-Tab metadata file
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
The Creative Commons Public Domain Dedication waiver http://creativecommons.org/publicdomain/zero/1.0/ applies to the metadata files associated with this article.
About this article
Cite this article
Liu, C., Wang, M., Wei, X. et al. An ATAC-seq atlas of chromatin accessibility in mouse tissues. Sci Data 6, 65 (2019). https://doi.org/10.1038/s41597-019-0071-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-019-0071-0
This article is cited by
-
Multi-omic characterization of allele-specific regulatory variation in hybrid pigs
Nature Communications (2024)
-
A spatiotemporal atlas of mouse liver homeostasis and regeneration
Nature Genetics (2024)
-
Profiling the immune epigenome across global cattle breeds
Genome Biology (2023)
-
Dissecting cell identity via network inference and in silico gene perturbation
Nature (2023)
-
Inferring mammalian tissue-specific regulatory conservation by predicting tissue-specific differences in open chromatin
BMC Genomics (2022)