Abstract
Poly(ADP-ribose) polymerase 1 (PARP1) activity is regulated by its co-factor histone poly(ADP-ribosylation) factor 1 (HPF1). The complex formed by HPF1 and PARP1 catalyzes ADP-ribosylation of serine residues of proteins near DNA breaks, mainly PARP1 and histones. However, the effect of HPF1 on DNA repair regulated by PARP1 remains unclear. Here, we show that HPF1 controls prolonged histone ADP-ribosylation in the vicinity of the DNA breaks by regulating both the number and length of ADP-ribose chains. Furthermore, we demonstrate that HPF1-dependent histone ADP-ribosylation triggers the rapid unfolding of chromatin, facilitating access to DNA at sites of damage. This process promotes the assembly of both the homologous recombination and non-homologous end joining repair machineries. Altogether, our data highlight the key roles played by the PARP1/HPF1 complex in regulating ADP-ribosylation signaling as well as the conformation of damaged chromatin at early stages of the DNA damage response.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The data sets generated and analyzed during the current study are not publicly available, as the large amount of imaging data could not be uploaded to a repository, but are available from the corresponding author upon request. Source data are provided with this paper.
Code availability
The MATLAB codes used in this work are available at https://github.com/sehuet/Smith-Zentout-image-processing.
References
Kraus, W. L. & Hottiger, M. O. PARP-1 and gene regulation: progress and puzzles. Mol. Aspects Med. 34, 1109–1123 (2013).
Chaudhuri, A. R. & Nussenzweig, A. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat. Rev. Mol. Cell Biol. 18, 610–621 (2017).
Eustermann, S. et al. Structural basis of detection and signaling of DNA single-strand breaks by human PARP-1. Mol. Cell 60, 742–754 (2015).
Ali, A. A. E. et al. The zinc-finger domains of PARP1 cooperate to recognize DNA strand breaks. Nat. Struct. Mol. Biol. 19, 685–692 (2012).
Langelier, M.-F., Planck, J. L., Roy, S. & Pascal, J. M. Structural basis for DNA damage–dependent poly(ADP-ribosyl)ation by human PARP-1. Science 336, 728–732 (2012).
Leidecker, O. et al. Serine is a new target residue for endogenous ADP-ribosylation on histones. Nat. Chem. Biol. 12, 998–1000 (2016).
Buch-Larsen, S. C. et al. Mapping physiological ADP-ribosylation using activated ion electron transfer dissociation. Cell Rep. 32, 108176 (2020).
Gibbs-Seymour, I., Fontana, P., Rack, J. G. M. & Ahel, I. HPF1/C4orf27 Is a PARP-1-interacting protein that regulates PARP-1 ADP-ribosylation activity. Mol. Cell 62, 432–442 (2016).
Suskiewicz, M. J. et al. HPF1 completes the PARP active site for DNA damage-induced ADP-ribosylation. Nature 579, 598–602 (2020).
Palazzo, L. et al. Serine is the major residue for ADP-ribosylation upon DNA damage. eLife 7, e34334 (2018).
Bonfiglio, J. J. et al. Serine ADP-ribosylation depends on HPF1. Mol. Cell 65, 932–940.e6 (2017).
Hendriks, I. A. et al. The regulatory landscape of the human HPF1- and ARH3-dependent ADP-ribosylome. Nat. Commun. 12, 5893 (2021).
Rudolph, J., Roberts, G., Muthurajan, U. M. & Luger, K. HPF1 and nucleosomes mediate a dramatic switch in activity of PARP1 from polymerase to hydrolase. eLife 10, e65773 (2021).
Sun, F.-H. et al. HPF1 remodels the active site of PARP1 to enable the serine ADP-ribosylation of histones. Nat. Commun. 12, 1028 (2021).
Mahadevan, J. et al. Q-FADD: a mechanistic approach for modeling the accumulation of proteins at sites of DNA damage. Biophys. J. 116, 2224–2233 (2019).
Langelier, M.-F., Billur, R., Sverzhinsky, A., Black, B. E. & Pascal, J. M. HPF1 dynamically controls the PARP1/2 balance between initiating and elongating ADP-ribose modifications. Nat. Commun. 12, 6675 (2021).
Prokhorova, E. et al. Serine-linked PARP1 auto-modification controls PARP inhibitor response. Nat. Commun. 12, 4055 (2021).
Juhász, S. et al. The chromatin remodeler ALC1 underlies resistance to PARP inhibitor treatment. Sci. Adv. 6, eabb8626 (2020).
Shao, Z. et al. Clinical PARP inhibitors do not abrogate PARP1 exchange at DNA damage sites in vivo. Nucleic Acids Res. 48, 9694–9709 (2020).
Gibson, B. A., Conrad, L. B., Huang, D. & Kraus, W. L. Generation and characterization of recombinant antibody-like ADP-ribose binding proteins. Biochemistry 56, 6305–6316 (2017).
Timinszky, G. et al. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat. Struct. Mol. Biol. 16, 923–929 (2009).
Wang, Z. et al. Recognition of the iso-ADP-ribose moiety in poly(ADP-ribose) by WWE domains suggests a general mechanism for poly(ADP-ribosyl)ation-dependent ubiquitination. Genes Dev. 26, 235–240 (2012).
Smith, R. et al. Poly(ADP-ribose)-dependent chromatin unfolding facilitates the association of DNA-binding proteins with DNA at sites of damage. Nucleic Acids Res. 47, 11250–11267 (2019).
Sellou, H. et al. The poly(ADP-ribose)-dependent chromatin remodeler Alc1 induces local chromatin relaxation upon DNA damage. Mol. Biol. Cell 27, 3791–3799 (2016).
Rother, M. B. et al. CHD7 and 53BP1 regulate distinct pathways for the re-ligation of DNA double-strand breaks. Nat. Commun. 11, 5775 (2020).
Smith, R., Sellou, H., Chapuis, C., Huet, S. & Timinszky, G. CHD3 and CHD4 recruitment and chromatin remodeling activity at DNA breaks is promoted by early poly(ADP-ribose)-dependent chromatin relaxation. Nucleic Acids Res. 46, 6087–6098 (2018).
Prokhorova, E. et al. Unrestrained poly-ADP-ribosylation provides insights into chromatin regulation and human disease. Mol. Cell 81, 2640–2655.e8 (2021).
Singh, J. K. et al. Zinc finger protein ZNF384 is an adaptor of Ku to DNA during classical non-homologous end-joining. Nat. Commun. 12, 6560 (2021).
Leung, J. W. C. et al. ZMYM3 regulates BRCA1 localization at damaged chromatin to promote DNA repair. Genes Dev. 31, 260–274 (2017).
Moison, C. et al. Zinc finger protein E4F1 cooperates with PARP-1 and BRG1 to promote DNA double-strand break repair. Proc. Natl Acad. Sci. USA 118, e2019408118 (2021).
Grundy, G. J. et al. APLF promotes the assembly and activity of non-homologous end joining protein complexes. EMBO J. 32, 112–125 (2013).
Liu, C., Vyas, A., Kassab, M. A., Singh, A. K. & Yu, X. The role of poly ADP-ribosylation in the first wave of DNA damage response. Nucleic Acids Res. 45, 8129–8141 (2017).
Kurgina, T. A. et al. Dual function of HPF1 in the modulation of PARP1 and PARP2 activities. Commun. Biol. 4, 1259 (2021).
Bonfiglio, J. J. et al. An HPF1/PARP1-based chemical biology strategy for exploring ADP-ribosylation. Cell 183, 1086–1102.e23 (2020).
Barkauskaite, E., Jankevicius, G., Ladurner, A. G., Ahel, I. & Timinszky, G. The recognition and removal of cellular poly(ADP-ribose) signals. FEBS J. 280, 3491–3507 (2013).
Murai, J. et al. Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res. 72, 5588–5599 (2012).
Poirier, G. G., de Murcia, G., Jongstra-Bilen, J., Niedergang, C. & Mandel, P. Poly(ADP-ribosyl)ation of polynucleosomes causes relaxation of chromatin structure. Proc. Natl Acad. Sci. USA 79, 3423–3427 (1982).
Hananya, N., Daley, S. K., Bagert, J. D. & Muir, T. W. Synthesis of ADP-ribosylated histones reveals site-specific impacts on chromatin structure and function. J. Am. Chem. Soc. 143, 10847–10852 (2021).
de Murcia, G. et al. Modulation of chromatin superstructure induced by poly(ADP-ribose) synthesis and degradation. J. Biol. Chem. 261, 7011–7017 (1986).
Luijsterburg, M. S. et al. PARP1 links CHD2-mediated chromatin expansion and H3.3 deposition to DNA repair by non-homologous end-joining. Mol. Cell 61, 547–562 (2016).
Bacic, L. et al. Structure and dynamics of the chromatin remodeler ALC1 bound to a PARylated nucleosome. eLife 10, e71420 (2021).
Mohapatra, J. et al. Serine ADP-ribosylation marks nucleosomes for ALC1-dependent chromatin remodeling. eLife 10, e71502 (2021).
Tulin, A. & Spradling, A. Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science 299, 560–562 (2003).
Mehrotra, P. V. et al. DNA repair factor APLF is a histone chaperone. Mol. Cell 41, 46–55 (2011).
Beaudouin, J., Mora-Bermúdez, F., Klee, T., Daigle, N. & Ellenberg, J. Dissecting the contribution of diffusion and interactions to the mobility of nuclear proteins. Biophys. J. 90, 1878–1894 (2006).
Polo, S. E., Kaidi, A., Baskcomb, L., Galanty, Y. & Jackson, S. P. Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. EMBO J. 29, 3130–3139 (2010).
Richardson, C., Moynahan, M. E. & Jasin, M. Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations. Genes Dev. 12, 3831–3842 (1998).
Densham, R. M. et al. Human BRCA1–BARD1 ubiquitin ligase activity counteracts chromatin barriers to DNA resection. Nat. Struct. Mol. Biol. 23, 647–655 (2016).
Britton, S., Coates, J. & Jackson, S. P. A new method for high-resolution imaging of Ku foci to decipher mechanisms of DNA double-strand break repair. J. Cell Biol. 202, 579–595 (2013).
Czarna, A. et al. Structures of Drosophila cryptochrome and mouse cryptochrome1 provide insight into circadian function. Cell 153, 1394–1405 (2013).
Gunn, A. & Stark, J. M. I-SceI-based assays to examine distinct repair outcomes of mammalian chromosomal double strand breaks. Methods Mol. Biol. 920, 379–391 (2012).
Tang, J. et al. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat. Struct. Mol. Biol. 20, 317–325 (2013).
Platani, M., Goldberg, I., Lamond, A. I. & Swedlow, J. R. Cajal body dynamics and association with chromatin are ATP-dependent. Nat. Cell Biol. 4, 502–508 (2002).
Wachsmuth, M. et al. High-throughput fluorescence correlation spectroscopy enables analysis of proteome dynamics in living cells. Nat. Biotechnol. 33, 384–389 (2015).
Haralick, R. M., Shanmugam, K. & Dinstein, I. Textural features for image classification. IEEE Trans. Syst. Man Cybern. SMC-3, 610–621 (1973).
Acknowledgements
We thank the Microscopy-Rennes Imaging Center (BIOSIT, Université Rennes 1), a member of the national infrastructure France-BioImaging supported by the French National Research Agency (ANR-10-INBS-04), for providing access to the imaging setups, as well as S. Dutertre and X. Pinson for technical assistance on the microscopes. We also thank the Cytométrie en flux et tri cellulaire (BIOSIT, Université Rennes 1), specifically L. Deleurme and A. Aimé for technical assistance with flow cytometry. We also thank M. Suskiewicz for thoughtful discussions and generously sharing the HPF1 plasmid DNA. For this work, S.H.’s group received financial support from the Agence Nationale de la Recherche (PRC-2018 REPAIRCHROM), the Institut National du Cancer (PLBIO-2019) and the Institut Universitaire de France. R.S. is supported by the Fondation ARC pour la recherche sur le cancer (PDF20181208405). The work in G.T.’s laboratory was supported by the Hungarian Academy of Sciences (LP2017-11/2017) and the National Research Development and Innovation Office (K128239). The work in I.A.’s laboratory is supported by the Wellcome Trust (210634 and 223107), Biotechnology and Biological Sciences Research Council (BB/R007195/1), Ovarian Cancer Research Alliance (813369) and Cancer Research United Kingdom (C35050/A22284). H.v.A.’s laboratory is financially supported by a NWO-VICI grant (VI.C.182.052) from the Dutch Research Council (NWO).
Author information
Authors and Affiliations
Contributions
R.S., G.T. and S.H. conceived the project with input from all authors. R.S., S.Z. and S.H. performed live-cell imaging and analyzed the imaging data. R.S., S.Z. and A.M performed ADPr western blots. N.B. performed colony formation assays. S.Z. performed the DNA repair assay and generated PARPKO stable cell lines. C.C. and R.S. generated DNA constructs. F.F.Z. and I.A. generated U2OS Flp-In/T-REx HPF1KO cells. M.R. and H.v.A. performed PLA and FokI assays. R.S., G.T. and S.H. wrote the paper with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Structural & Molecular Biology thanks Karolin Luger, Kumar Somyajit and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Beth Moorefield and Carolina Perdigoto, in collaboration with the Nature Structural & Molecular Biology team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 HPF1 recruits to sites of DNA damage.
(a) Representative images of HPF1 recruitment to sites of DNA damage in the presence or absence of PARPi. The top panels show endogenous HPF1 recruitment while the bottom panels show GFP-HPF1 recruitment. Red arrows indicate sites of laser microirradiation with 405 nm. For HPF1 immunostaining, cells were fixed immediately after irradiation. For the GFP-HPF1 expressing cells, images were taken 120 s post-irradiation. Scale bar, 5 µm. (b) Analysis of PARP1 and HPF1 retention on chromatin after H2O2 treatment by chromatin fractionation and western blotting. Soluble and chromatin fractions are shown. H2B is used as a control demonstrating effective fractionation. (c–f) Proximity Ligation Assay (PLA) of GFP and γH2AX in U2OS Flp-In/T-Rex cells expressing doxycycline-inducible YFP-HPF1 (c,d) or YFP-PARP1 (e,f) irradiated or not with 2 Gy of ionizing radiation (IR). PLA signal was quantified as the number of PLA spots per nuclei 1 h after DSB induction. As a negative control only one primary antibody (GFP-only or γH2AX-only) was used. Representative images from >50 cells of a representative experiment from 2 independent replicates are shown. Scale bar, 5 µm (d) Quantification of c. Data from d are a representative of 3 independent replicates where data were collected from 49–111 cells per condition. (f) Quantification of e. Data from f are a representative of 3 independent replicates where data were collected from 88–119 cells per condition. (g) Schematic illustrating calculation of residual protein accumulation. Initially, the tmax and t1/2 are determined from the mean PARP1 WT recruitment curve where tmax is the time where PARP1 WT recruitment is maximal, and t1/2 is the time post-irradiaion where the recruitment has fallen to 50% PARP1 WT recruitment (Left). Then for each recruitment curve of PARP1 (WT or mutant) and HPF1, maximal recruitment was normalised to tmax and the residual accumulation was taken at t1/2 (Right).
Extended Data Fig. 2 HPF1 recruitment to sites of damage relies on interaction with the C-terminus of PARP1.
(a, b) Western blot analysis (a) and representative confocal images (b) of YFP-HPF1 expression levels in U2OS Flp-In/T-Rex HPF1KOcells after doxycycline induced expression. Transient transfection with YFP-HPF1 was also used to further boost HPF1 expression in the presence of 25 ng/mL Doxycycline. Images were taken 30 s post 405-nm laser irradiation. Scale bar, 10 μm. (c) Quantification of the mean nuclear fluorescence intensity of cells from b. Fluorescence level is normalised to 5 ng/mL (endogenous levels of HPF1 expression according to a). (d, e) Mean (d) or normalized (e) recruitment kinetics of YFP-HPF1 after 405-nm laser irradiation in U2OS Flp-In/T-Rex HPF1KOcells with different levels of doxycycline induction. Individual recruitment curves from d were normalised to the peak HPF1 recruitment levels. Data from b-e are a representative of 2 independent replicates where data were collected from 12–18 cells per condition. (f) Recruitment kinetics of GFP-HPF1 in PARP1KO or PARP1/HPF1 double knockout cells expressing mCherry-PARP1 WT. Data from f are a representative of 2 independent replicates where data were collected from 10–12 cells per condition. (g) Immunoblots of whole-cell extract from U2OS WT, PARP1KO, HPF1KO and PARP1/HPF1 double knockout cells. (h) Recruitment kinetics of GFP-HPF1 to sites of DNA damage induced by 405-nm laser irradiation in WT or PARP1KO cells expressing WT N-terminally (mCh-PARP1) and C-terminally tagged PARP1 (PARP1-mCh). Data from h are a representative of 3 independent replicates where data were collected from 12–16 cells per condition. (i) Schematic representation of PAR-3H assay. (j, k) Representative confocal images (j) or quantification (k) of mCherry tagged PARP1 WT, PARP1 3SA or PARP1 E988K recruitment to YFP tagged macrodomain of mH2A1.1 tethered to LacO, Pre or 30 s post-irradiation. Inset, pseudocolored according to the look-up table displayed, shows the magnified LacO array. Scale bar, 5 µm. Data from k are a representative of 3 independent replicates where data were collected from 13–15 cells per condition. (l, m) Normalised recruitment kinetics of mCherry tagged PARP1 (WT, 3SA or E988K) (l) and GFP-HPF1 (m) expressed in PARP1KO cells from Fig. 1f–h. Data from l-m are a representative of 3 independent replicates where data were collected from 13–18 cells per condition.
Extended Data Fig. 3 HPF1 regulates ADP-ribosylation signaling at sites of DNA damage.
(a) Recruitment kinetics of GFP-macrodomain of mH2A1.1 at sites of DNA damage induced by 405 nm laser irradiation, in U2OS PARP1KO cells complemented or not with mCherry-PARP1 WT, PARP1 3SA or PARP1 LW/AA. (b) Quantification of mean recruitment intensity of GFP-macrodomain of mH2A1.1 at sites of DNA damage 200 s post-irradiation in PARP1KO complemented or not with mCherry-PARP1 WT or PARP1 LW/AA mutants. ∅ denotes no plasmid expression. Data from a,b are a representative of 3 independent replicates where data were collected from 10–17 cells per condition.
Extended Data Fig. 4 HPF1 promotes chromatin relaxation at sites of DNA damage.
(a) Chromatin relaxation in U2OS WT and HPF1KO cells at 60 s post irradiation with 800 nm laser. Data from a are a representative of 3 independent replicates where data were collected from 16–19 cells per condition. (b) Chromatin relaxation in U2OS WT and PARP1KO cells at 120 s post-irradiation at 405 nm. Cells are complemented or not with C-terminally-tagged PARP1-mCherry. ∅ denotes no plasmid expression. Data from b are a representative of 3 independent replicates where data were collected from 10–17 cells per condition. (c) Recruitment kinetics of mCherry-tagged HPF1 WT and the point mutants D283A and E284A at sites of DNA damage induced by 405 nm laser irradiation in U2OS HPF1KO cells. Data from c are a representative of 3 independent replicates where data were collected from 10–14 cells per condition. (d) Confocal images of U2OS WT and HPF1KO nuclei stained with Hoechst. Scale bar, 5 μm. Images are coloured according to the lookup table below the cells. (e) The chromatin pattern in Hoechst-stained cells was characterised by the image contrast, calculated as the mean squared intensity difference of pixels separated by a distance of 7 pixels. Data from e is a representative of 2 independent replicates where data were collected from 135–190 cells per condition. (f) Chromatin relaxation and HPF1 recruitment kinetics at sites of DNA damage induced by 405 nm laser irradiation in WT cells overexpressing mCherry-HPF1. Data shown is mean ± SEM. Data from f are a representative of 3 independent replicates where data were collected from 20 cells per condition.
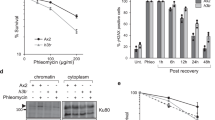
Extended Data Fig. 5 HPF1-dependent chromatin relaxation relies on trans ADP-ribosylation of histones rather than PARP1 auto-modification.
(a) Western blot displaying ADPr signals, stained with a pan-ADPr antibody, in WT and HPF1KO cells expressing HPF1 WT or HPF1 E284A and treated or not with H2O2. H2B and Tubulin were used as loading controls. (b) Western blot displaying ADPr signals, stained with a pan-ADPr antibody, in U2OS WT, PARP1KO and PARP1/HPF1 double knockout cells expressing mCherry tagged PARP1 WT, PARP1 3SA or PARP1 LW/AA and treated or not with H2O2. H3 and Tubulin were used as loading controls. (c) Representative images of the recruitment of mCherry-tagged HPF1 WT or R239A to sites DNA damage induced by 405 nm laser irradiation in U2OS HPF1KO cells. Scale bar, 5 μm. (d) Recruitment kinetics of mCherry-tagged HPF1 WT or HPF1 R239A mutant at sites of DNA damage in U2OS HPF1KO cells. Data from d are a representative of 3 independent replicates where data were collected from 15–16 cells per condition. (e) Western blot displaying ADPr signals, stained with a pan-ADPr antibody, in WT cells overexpressing YFP-HPF1 or not with or without ATP depletion (ATPi) after H2O2 damage. H3 and Tubulin were used as loading controls. (f) Chromatin relaxation 120 s post irradiation at 405 nm in U2OS WT or ALC1KO cells overexpressing HPF1 or not. Data from f are a representative of 3 independent replicates where data were collected from 16–18 cells per condition. (g) Western blot analysis confirming knockout status of ALC1KO cells. (h) Chromatin relaxation 120 s post DNA damage in U2OS WT or CHD7KO cells overexpressing HPF1 or not. Data from h are a representative of 3 independent replicates where data were collected from 15–18 cells per condition. (i) Western blot analysis confirming knockout status of CHD7KO cells.
Extended Data Fig. 6 HPF1 regulates the recruitment of DNA-binding repair factors to sites of damage.
(a) Immunoflurosecence showing depletion of HPF1 in ZNF384KO cells. Hoechst staining shows nuclei. Scale bar, 10 μm. (b-d) Representative confocal images (b), recruitment kinetics (c), and mean recruitment intensity at 200 s post-irradiation (d) of GFP-E4F1 at sites DNA damage induced by 405 nm irradiation in U2OS WT and HPF1KO cells treated or not with PARPi. Confocal images are 200 s post-irradiation. Scale bar, 5 μm. Recruitment kinetic curves show the mean ± SEM Data from c,d are a representative of 3 independent replicates where data were collected from 11–13 cells per condition. (e, f) Representative confocal images (e) and recruitment kinetics (f) of GFP-CHD4 at sites DNA damage induced by 405 nm irradiation in U2OS WT, PARP1KO and HPF1KO cells. Confocal images are 200 s post-irradiation. Scale bar, 5 μm. Recruitment kinetic curves show the mean ± SEM. Data from f are a representative of 3 independent replicates where data were collected from 13–22 cells per condition. (g, h) Representative confocal images (g) and recruitment kinetics (h) of GFP-CHD7 at sites DNA damage induced by 405 nm irradiation in U2OS WT, PARP1KO and HPF1KO cells. Confocal images are 200 s post-irradiation. Scale bar, 5 μm. Recruitment kinetic curves show the mean ± SEM. Data from h are a representative of 3 independent replicates where data were collected from 13–16 cells per condition.
Extended Data Fig. 7 HPF1-dependent recruitment of DNA-binding repair factors to sites of damage is promoted by histone ADP-ribosylation.
(a–c) Representative confocal images (a), recruitment kinetics (b), and mean recruitment intensity at 200 s post-irradiation (c) of GFP-E4F1 at sites DNA damage induced by 405 nm irradiation in U2OS PARP1KO cells complemented or not with mCherry-PARP1 WT, PARP1 3SA or PARP1 LW/AA. Confocal images are 200 s post-irradiation. Scale bar, 5 μm. Recruitment kinetic curves show the mean±SEM. Data from b,c are a representative of 3 independent replicates where data were collected from 12–16 cells per condition. (d-f) Representative confocal images (d), recruitment kinetics (e), and mean recruitment intensity at 200 s post-irradiation (f) of GFP-CHD4 at sites DNA damage induced by 405 nm irradiation in U2OS PARP1KO or PARP1/HPF1 double knockout cells complemented or not with mCherry-PARP1 WT, PARP1 3SA or PARP1 LW/AA mutants. ∅ denotes no plasmid expression. Confocal images are 200 s post-irradiation. Scale bar, 5 μm. Recruitment kinetic curves show the mean ± SEM. Data from e,f are a representative of 3 independent replicates where data were collected from 10–13 cells per condition. (g-i) Representative confocal images (g), recruitment kinetics (h), and mean recruitment intensity at 200 s post-irradiation (i) of GFP-CHD7 at sites DNA damage induced by 405 nm irradiation in U2OS PARP1KO or PARP1/HPF1 double knockout cells complemented or not with mCherry-PARP1 WT, PARP1 3SA or PARP1 LW/AA mutants. ∅ denotes no plasmid expression. Confocal images are 200 s post-irradiation. Scale bar, 5 μm. Recruitment kinetic curves show the mean ± SEM. Data from h,i are a representative of 3 independent replicates where data were collected from 10–12 cells per condition.
Extended Data Fig. 8 HPF1 promotes efficient DNA repair.
(a, b) Representative images of clonogenic assay (a) and cell survival curves (b) for WT and PARP1KO cells upon continuous camptothecin treatment. Data are a representative of 3 independent replicates. (c) Western blot displaying ADPr signals, stained with a pan-ADPr antibody, in U2OS WT, PARP1KO and PARP1KO cells stably expressing GFP tagged PARP1 WT, PARP1 3SA or PARP1 LW/AA and treated or not with H2O2. H3 and Tubulin were used as loading controls. (d) Gating strategy used to analyze DNA repair efficiency. Gate A was used to select living cells: SSC-A scatter against FSC-A. Gate B was used to remove of doublets: FSC-H scatter against FSC-A. The mCherry positive population was selected in Gate C where mCherry positive cells had above background PE-CF594-A signal. The GFP-positive population (FITC-A) was selected and counted from mCherry positive cells in Gate C (final panel). (e) Schematic representation of the HR reporter assay (DR). After cleavage with I-SceI, the double-strand-breaks repaired by HR results in GFP expression. (f) Representative immunoblots showing the knockdown efficiency of BRCA2 and HPF1 in DR cells. Actin is used as a loading control. (g) Schematic representation of the NHEJ reporter assay (EJ5). Double cleavage by I-SceI removes the Puro cassette and the repair of the double-strand-break by NHEJ allows GFP expression. (h) Representative immunoblots showing the knockdown efficiency of XRCC4 and HPF1 in EJ5 cells. Actin is used as a loading control.
Supplementary information
Supplementary Information
Supplementary Tables 1–3.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 1
Unprocessed western blots
Source Data Fig. 2
Statistical source data.
Source Data Fig. 2
Unprocessed western blots.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 4
Unprocessed western blots.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 5
Unprocessed western blots.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 7
Statistical source data.
Source Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 1
Unprocessed western blots.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 2
Unprocessed western blots.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 5
Unprocessed western blots.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 8
Unprocessed western blots.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Smith, R., Zentout, S., Rother, M. et al. HPF1-dependent histone ADP-ribosylation triggers chromatin relaxation to promote the recruitment of repair factors at sites of DNA damage. Nat Struct Mol Biol 30, 678–691 (2023). https://doi.org/10.1038/s41594-023-00977-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-023-00977-x