Abstract
Simian immunodeficiency viruses (SIVs) are lentiviruses that naturally infect non-human primates of African origin and seeded cross-species transmissions of HIV-1 and HIV-2. Here we report prefusion stabilization and cryo-EM structures of soluble envelope (Env) trimers from rhesus macaque SIV (SIVmac) in complex with neutralizing antibodies. These structures provide residue-level definition for SIV-specific disulfide-bonded variable loops (V1 and V2), which we used to delineate variable-loop coverage of the Env trimer. The defined variable loops enabled us to investigate assembled Env-glycan shields throughout SIV, which we found to comprise both N- and O-linked glycans, the latter emanating from V1 inserts, which bound the O-link-specific lectin jacalin. We also investigated in situ SIVmac-Env trimers on virions, determining cryo-electron tomography structures at subnanometer resolutions for an antibody-bound complex and a ligand-free state. Collectively, these structures define the prefusion-closed structure of the SIV-Env trimer and delineate variable-loop and glycan-shielding mechanisms of immune evasion conserved throughout SIV evolution.
Similar content being viewed by others
Main
Simian immunodeficiency viruses (SIVs) infect over 40 species of African monkeys, chimpanzees and gorillas1,2 and are well-studied examples of cross-species infection and adaptation3,4,5,6. Although rapid viral sequence evolution complicates dating of cross-species transmission, analysis of SIV from monkeys on Bioko Island suggests SIV to have been present in African primates for more than 32,000 years7, and possibly much longer8. Multiple cross-species transmissions of SIV from chimpanzee (SIVCPZ) to humans have resulted in groups M, N and O of HIV-1, with group M having crossed ~100 years ago to seed the HIV-1 pandemic9,10, which has killed an estimated 27–48 million and currently infects 38 million humans11. Eight distinct crossovers of SIV from sooty mangabeys (SIVSMM) have resulted in groups A–H of HIV-2, although only groups A and B are found widely12,13.
Entry of these immunodeficiency-causing viruses into target cells depends on the viral envelope (Env) spike, which binds CD4 and its coreceptor (reviewed in refs. 14,15). The Env spike is a trimer of hetero-dimers comprising two heavily glycosylated proteins: gp120, containing the receptor-binding sites, and gp41, containing the virus-fusion machinery. For both SIV and HIV, Env is the sole target of neutralizing antibodies as it is the only viral antigen on the surface of infectious virus particles, and as such it has evolved multiple mechanisms of immune evasion, including sequence diversity10,16, conformational masking17, glycan shielding18 and low-spike density19,20. Evolutionary pressure from both viral and host factors in the transmission to, and adaptation in, new hosts has resulted in Env insertions, deletions and differing glycan usage throughout SIV. These cannot be accurately modeled due to SIV-specific disulfide-bonded loops and a lack of structural details from SIVs other than the direct HIV-1 group M progenitor, SIVCPZ (ref. 21), which lacks loops of comparable size and composition. Direct application to SIV of trimer-stabilizing mutations effective for the HIV-1 Env22 succeeds with the more closely related chimpanzee SIVcpz Env21, but does not result in homogeneous trimers of SIVmac, the virus used to infect rhesus macaques as the most commonly studied pathogenic animal model for HIV. In this Article we use a structure-based approach to stabilize soluble recombinantly expressed SIVmac Env trimers, for which we determined cryo-EM structures in complexes with antibodies ITS92.02 and PGT14523. We delineate variable-loop shielding of the SIVmac Env trimer along with its glycan shielding. Finally, we determine in situ structures of the SIV Env trimer on virions, both ligand-free and in complex with antibody ITS90.0324, which fully neutralizes SIVmac239. Because SIVmac is the primary animal-model pathogen for HIV-1, the structural details provided here, along with the development of a soluble SIV Env trimer, should enable a wide array of studies, as demonstrated with HIV-1.
Results
SIV Env trimer stabilization and structure determination
HIV-1 and SIVmac Envs share ~30% sequence identity. However, the cysteine mutations at positions 501 and 605 (SOS) with a proline mutation at 559 (IP) used in combination (SOSIP22) that stabilize the prefusion conformation of HIV-1-Env trimers did not allow for the formation of a soluble prefusion-stabilized SIVmac-Env trimer (Extended Data Fig. 1a). With HIV-1 Env as the structural template, we designed 57 additional mutations and assessed their impact on the antigenicity of SIVmac-Env trimers in a 96-well transient-expression format. Although several candidate variants were tested with either SOS or IP mutations, most were primarily based on SOSIP constructs supplemented with additional disulfides, strain variation, cavity-filling mutations or proline insertions. We screened for mutants that showed improved binding to antibody ITS92.02, which neutralizes the SIV E660.CR54 strain but does not bind to the monomeric gp120 of that strain, suggesting it may prefer an Env conformation more closely related to that found on the virion surface. The best antigenic properties were observed in a design we named SOS-2P, which contained SOSIP alterations supplemented with a second proline mutation, T569P (HXB2 numbering; Fig. 1a); this mutation had previously been identified as a potential stabilizing mutation for HIV-1 Env25, highlighting the broad applicability of prefusion di-proline stabilization26,27.
a, Left: designs of stabilized soluble SIV trimers based on HIV-1 structures were assessed through antigenic screening of a 96-well plate. Middle: antigenicity for the designs, shown as a heatmap, with each designed construct as a row and each antibody as a column, with ITS92.02 on the rightmost column (Extended Data Fig. 1b). Right: introduction of a proline mutation at 569, in addition to the proline mutation of SOSIP at 559, showed the best binding to ITS92.02. b, SIVCR54 SOS-2P with ITS92.02 cryo-EM density. Density at the apex is less well-defined than the core and gp41 regions. c, The atomic model of the structure is shown in cartoon format. d, Density at the ITS92.02 epitope, shown with the model. e, The ITS92.02 epitope is highlighted in red and shown in the bound-prefusion conformation (looking up from the membrane) of SIVE660.CR54 and the postfusion conformation (side view; PDB 1QBZ) of SIVmac239. f, The density of the SIV trimer (left) did not allow for unambiguous model-building of the apex region, including the V1V2 and V3 domains, which are highlighted in HIV-1 (right; PDB 4TVP) in blue and orange, respectively.
We purified the SOS-2P-stabilized SIV-Env trimer from the E660.CR54 strain in complex with ITS92.02 Fab (Extended Data Fig. 1b) and obtained a cryo-EM structure with a global FSC0.143 resolution of 4.3 Å. The ITS92.02 Fab was revealed to be specific for an epitope in the prefusion gp41 conformation (Fig. 1c–e and Extended Data Figs. 2f,g) that is not retained in the postfusion conformation28 and is sequence-specific to SIV strain E660.CR54. Although densities for the Fv portion of Fab and for gp41 were well-resolved, with local resolutions of ~3.5 Å, density proximal to the trimer apex was poor with local resolution of 8 Å or worse, leaving it unsuitable for insight into the V1V2 and V3 regions (Fig. 1d,f, Extended Data Figs. 1c and 2a–e and Table 1).
To stabilize the trimer apex, we used apex-directed antibody PGT14523,29,30,31, which we observed neutralizes SIVmac251.30 (Fig. 2a,b) and has been found to bind a membrane-bound K169T mutant32 of the Env trimer of SIVmac23933,34, a difficult-to-neutralize, tier-3 SIV strain. The mac251.30 Env has a Thr at residue 16933, suggesting this residue to play an important role in PGT145 binding. Co-expression of the SIVmac239 SOS-2P trimer with the K169T mutation in complex with PGT145 paired with on-column purification enabled us to obtain a stabilized SIV Env trimer suitable for cryo-EM. We used the Fab PGT145-bound Env complex to determine the cryo-EM structure of the SIVmac239 trimer to a resolution of 4.1 Å (Fig. 2c, Extended Data Fig. 3 and Table 1), revealing a well-resolved trimer apex (Fig. 2d).
a, PGT145 neutralization curves are shown for three SIV viruses. WT SIVmac251.30, which has a native Thr at position 169, is neutralized. The error bars indicate the s.d. of duplicate measurements. b, The 169S/T mutation at the trimer apex required for PGT145 to bind and neutralize SIV is shown in red. c, Cryo-EM density at 4.1 Å, shown for SIVmac239 SOS-2P trimer with the K169T mutation, in complex with PGT145. d, Density at the apex allowed unambiguous model-building for the trimer and PGT145 CDR H3. e, The overall structure of the SIV Env trimer looking down the three-fold axis shows the three-blade propeller. gp120 and gp41 for each protomer are colored individually and labeled. f, Cα alignment of SIVmac239 protomer with HIV-1 (PDB 4TVP) and SIVcpz (PDB 6OHY). g, Modeling the native Lys at position 169 shows clashes with residues R100a and F100d of the PGT145 CDR H3, which are not seen in the K169T mutant structure.
Structure of a prefusion-closed SIV Env trimer
Overall, the SIVmac239 Env trimer formed a three-blade propeller structure (Fig. 2e), which—discounting inserts and deletions—aligned closely with the structure of the HIV-1 Env with a root-mean-square deviation (r.m.s.d.) of 2.3 Å over 1,302 Cα atoms, with the SIVcpz Env with a r.m.s.d. of 2.3 Å over 1,100 Cα atoms (Fig. 2f). The full-length gp120 of the trimer aligned with a r.m.s.d. of 0.9 Å to the core of SIVmac239 we previously determined, with minor differences observed in the glycan shield for common sequons in the structures (Extended Data Fig. 4a). A few typically disordered regions in HIV-1, including the V4 loop of gp120 and α6-helix of gp41, were ordered in the SIV structure, but the overall secondary and tertiary structures were conserved (Extended Data Fig. 4b). Most of the larger structural deviations were in variable loops and glycan shield. Modeling the native Lys at position 169 indicated a clash with the heavy chain third complementarity-determining region (CDR H3) of PGT145, explaining the necessity for a Thr mutation at this position enabling binding of this HIV-1 broadly neutralizing antibody (Fig. 2g).
Outside of the variable loops, large deviations also occurred at residues 54–74 in the gp120 subunit and residues 650–664 in the gp41 subunit (Extended Data Fig. 4c). Residues 54–74 are associated with two-point mutations, T55A and R57K, which dramatically impact the neutralization phenotype of SIV strains35. These residues formed a small hairpin. The equivalent region of HIV-1 encompasses a seven-residue insertion, 60–66HIV-1 (for clarity, the molecule is provided as a subscript when residue numbering is not SIV Env), which forms a short helix from residues 57–62HIV-1 followed by a loop; this region rearranges upon CD4 binding to form a helix in residues 63–74HIV-1 that stacks with the α7-helix of gp41 (Extended Data Fig. 5). Modeling suggested that the small hairpin formed in this region in SIVmac239 would clash with gp41 upon CD4 binding, if the region were to rearrange in a fashion analogous to HIV-1. Rhesus CD4 (rCD4) differs from human CD4, and HIV-1 entry with rCD4 is facilitated by mutation from Ser375HIV-1 or A281HIV-136,37. Residues 650–664 were located at the C terminus of the soluble construct, and their movement likely reflects mobility in this helix at the membrane-proximal face of the soluble Env trimer.
Extended SIV loops conceal analogous HIV-1 antibody epitopes
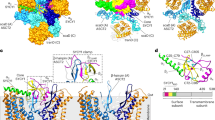
The most striking differences in the Env trimer structure between SIV and HIV-1 involved extended variable loops, particularly hypervariable loop V1. Following residue 139, SIVmac239 Env contains an insertion of 37 residues compared to that of HIV-1 BG505 Env, including an additional disulfide bond (Fig. 3a). Bioinformatics predicts this extended V1 loop to contain multiple potential O-linked glycosylation sites38 (POGs), and indeed V1 has been confirmed to contain O-linked glycans39,40,41 with soluble gp120 and virions produced by recombinant expression in multiple SIV strains including SIVmac239. Discounting this additional glycan coverage, the 37-residue extension in the V1 loop covered ~1,000 Å2 of the Env with a hypervariable protein surface. Much of the analogous surface in HIV-1 Env is shielded by glycosylation, suggesting glycosylation of these exposed apical regions may facilitate evasion from surveilling antibodies (Fig. 3b). Indeed, the hypervariable V1 loop-shielded surface included part of the coreceptor-binding site, which overlaps with 324GDIR327 and glycan N332HIV-1, part of the glycan-V3 supersite (Fig. 3c), which is one of the most commonly targeted sites in HIV-123,42,43,44. Shielding of this V3 region may influence the difference in sequence diversity arising from immune pressure in SIV compared to that in HIV-1.
a, Sequence alignment of HIV-1 and SIVmac239 V1V2 region, marking an extended SIV V1 loop with predicted O-linked glycosylation sites (POGs) in red. N-linked glycans are shown in gold and green for HIV-1 and SIV, respectively. b, Surface representation of HIV-1 (gray) with cartoon representation of SIVmac239 (magenta), highlighting the extended surface coverage by the SIV V1 and V4 loops, POGs are colored in red and V4 in blue. c, The surface region of the HIV-1 N332 (gold) V3-glycan epitope (cyan) is occluded by the SIV V1 loop. This region is marked by a cyan box in b. d, The SIVmac239 V1V2 region, highlighting the N-linked (green) and O-linked (red) glycosylation sites. The two disulfides not found in HIV-1 are shown in dark purple, with the resulting loops shown in thicker tube radius. e, The V4 loop of SIVmac239 is oriented towards the apex covering the region protected by glycans N363, N386 and N392 in HIV-1. The HIV-1 V4 is oriented away from the apex (gray, PDB 2B4C).
The SIVmac239 V2 loop is four residues shorter than that of BG505 and contained an additional disulfide bond proximal to the region of V2 that binds α4β7 in HIV-1 Env45. The additional disulfide stabilized the positioning of two N-linked glycans, at residues 185b and 187, which along with glycan N160, shielded much of the trimer apex (Fig. 3a,d). The V3 loop of SIVmac239, which was buried by the V1V2 domain, aligned closely with that of HIV-1. The V3 regions in both HIV-1 and SIV are more conserved than other variable loops, lacking true hypervariable sequence variation. The V4 loop of SIV was shorter than that of HIV-1 and does not extend away from the apex in a disordered conformation as in HIV-1, but folded and extended towards the V1V2 loops, where it abutted the O-glycosylated V1 insert (Fig. 3e). In general, the V4 region is structurally variable, and a residue-by-residue distance comparison confirmed large structural deviations between SIV and HIV-1 to occur in V4 (Extended Data Fig. 4c). The SIVmac239 V5 loop did not deviate substantially from that of HIV-1, and it contained an N-linked glycan, N463, corresponding to similar glycosylation observed in many HIV-1 strains. Thus, the disulfide-extended V1 and V2, along with the structurally mobile V4, increased shielding of the SIVmac239 apex and its periphery, whereas V3 and V5 appeared to more closely reprise their roles in HIV-1.
Evolutionarily conserved SIV features
Several features—observed in the SIVmac239 structure, but not in HIV-1—were conserved in SIV isolates that infect diverse primates (Fig. 4a). The V1 disulfide-bonded insert in SIVmac239 is found in HIV-2 and SIVsmm, whereas the additional V2-disulfide-bonded region is conserved across a number of divergent strains (Fig. 4a,b and Extended Data Fig. 6a,b). The structure of SIVmac239 defines the local conformations stabilized by the V1V2 disulfides and the regions they protect from, or expose to, the immune system. Sequence analysis revealed additional instances of cysteine pairs arising in multiple variable loops of diverse SIV branches46, suggesting specific evolutionary diversification arose from similar shielding strategies. Notably, only HIV-1 and SIVcpz lack V1 POGs, which are found in most SIVs (Fig. 4a,b). This corresponds with the much shorter V1 hypervariable loops found in those viruses (Fig. 4b). The presence of POGs did not appear to impact the total number of N-linked glycosylation sites (Fig. 4b). Additionally, an insertion near the α0-helix is present only in strains more closely related to HIV-1. Analysis of SIV sequence entropy (Fig. 4c), meanwhile, showed conservation at both the CD4-binding site and the apex near the trimer three-fold axis. The conserved use of CD4 as a receptor throughout SIV explains the conservation of the former, whereas the conservation of the latter provides an explanation for the ability of V2-apex-targeting antibodies such as PGT145 to neutralize broadly23,47. The SIVmac239 Env structure showed that the disulfide formed in V1 of SIVmac239 resides in a portion of the loop that deviated immensely from that of HIV-1BG505, whereas the disulfide in the V2 loop formed across two positions in adjacent β strands conserved in structures of both virus Envs (Fig. 4d,e). The seven-residue insertion at 60–66HIV-1 extended the region encompassed by the C54–C74 disulfide that rearranges upon CD4 binding (Fig. 4f and Extended Data Fig. 5c).
a, A phylogenetic tree of representative Env sequences from HIV-1, HIV-2 and SIV. Branches that conserve features such as the a0 insert and the V1 and V2 disulfides are highlighted. All branches except HIV-1 and SIVcpz are predicted to contain O-linked glycans. b, Amino acid lengths of the V1 (C131 and C157) and V2 (C183–C191) hypervariable regions and the number of N-linked sequons for the full Env. c, SIV sequence entropy, mapped to the surface of SIVmac239 with high sequence conservation in white and low sequence conservation in purple. d, The V1 hypervariable loops of HIV-1, SIVcpz and SIVmac239, overlayed with residues between the conserved disulfide bond at positions C131 and C157 following strand B and preceding strand C. The disulfide in SIVmac239 is shown in purple. e, The V2 regions from 179 to 191 are shown with an additional disulfide in SIVmac239 at positions 183 and 191, which are Gln and Tyr in HIV-1BG505 or Ala and Tyr in SIVcpzMT145K, respectively. BG505 structures are typically disordered in parts of this loop. f, The region proximal to α0 contains a seven-residue insert in HIV-1 and SIVcpz that is not observed in the majority of SIVs.
The SIV glycan shield
The cryo-EM density was of sufficient quality to observe details of the SIVmac239 glycan shield (Fig. 5a). Predicted N-linked glycosylation sites (PNGs) showed carbohydrate density for at least the protein-proximal N-acetyl glucosamine (GlcNac) for 25 of 26 sites, the exception being N232 (which directly follows the N229 sequon and is thus probably not fully occupied). In terms of N-linked glycan holes, SIVmac239 contains a glycan hole at residue position 238, which is glycosylated in 83% of SIV strains. This glycan hole exposes a region that is targeted by ITS90.03, an antibody that fully neutralizes the SIVmac239 strain24.
a, Cryo-EM density, with green regions corresponding to N-linked glycans and red regions O-linked glycans. b, Density, shown as mesh, surrounding V1 Thr residues predicted to have O-linked glycosylation. The resolution of the density in this region was not high enough to build with atomic-level certainty, but the level of excess density strongly suggested there were heterogeneously glycosylated residues in the loop. c, Full glycan shield for HIV-1 BG505 (gold; PDB 5FYL) and SIV mac239 (green and red), with some direct overlap (that is, N160, N197), some neighboring overlap (that is, SIV N229 with HIV-1 N241) and non-overlapping coverage (that is, SIV N47). d, Cryo-EM density of the PGT145-bound SIVmac239 trimer in complex with the O-linked glycan-binding jacalin is shown as transparent, with the atomic model fit and V1 loops colored in red. e, Modeled glycan shields based on SIV-HIV templates. Glycans are colored green if present in SIVmac239 and otherwise colored as in a and b by their initial appearance. Glycans that arise in hypervariable regions of V1, V2 or V4 or those shifted within two residues or less of a previously present glycan are not recolored. Sequence identity to SIVmac239 of the Env (31 to 664 by HXB2 numbering) is shown below, with other features of the related viruses, including alternate V1 disulfides (Extended Data Fig. 6) and POGS calculated from NetOGlyc-4.0 with a conservative cutoff score of over 0.75.
In terms of O-linked glycosylation, 12 POGs appear in the extended V1 loop (Fig. 3a). Although this loop has been experimentally confirmed to contain O-linked glycans39,40,41, flexibility of the loop and its glycans along with heterogeneity in O-linked glycosylation complicated the interpretation of electron density for modeling of individual glycan moieties. Nevertheless, density in the V1 region clearly indicated additional mass beyond that accounted for by loop amino acids (Fig. 5b). We note that, although the POG sequence signatures were experimentally confirmed as glycosylated here, O-linked glycosylation levels may differ in Envs expressed through natural infection.
To provide additional structural definition of O-linked glycans, we complexed the O-linked specific lectin, jacalin, to the PGT145-SIV Env trimer complex and determined its structure to a resolution of 7.6 Å. Density corresponding to the bound jacalin protruded from the V1 region (Fig. 5d), providing further confirmation of O-linked glycosylation. In terms of glycan shielding, the overall impact of O-linked glycan appeared to be low, with molecular dynamics simulations of 1–10 O-linked core-1 glycans revealing some impact to glycan coverage of the V3 region, while other epitope regions appeared to be mostly unaffected (Extended Data Fig. 7).
To extend our understanding from the defined glycan-shield structure of SIVmac239 to the divergent Env trimers throughout SIV, we carried out structural alignments of glycan, finding seven N-linked glycosylation sites to align directly with those in HIV-1 BG505 or JR-FL, while an additional six contained Asn ND2 nitrogen atom within ~5 Å of a matched HIV-1 glycan, and 12 were unmatched to HIV-1 (Fig. 5c and Extended Data Fig. 8). With V1- and V2-disulfide linked structures defined, we could model other SIV Env trimers based on our newly determined structures and known HIV-1 Env trimer structures. We modeled glycan shields of the Env trimers for various SIV strains, ranging in sequence identity to SIVmac239 from 35 to 83% (Fig. 3e). Despite substantial sequence and evolutionary divergence, we observed extensive glycan shields to be maintained throughout SIV. Overall, the SIV glycan shield, comprising both N-linked glycans and O-linked glycans (from V1), masked much of the Env-trimer surface, especially its apex.
In situ Env structures on SIVmac239 virions
We sought to confirm that the structure we determined of the soluble SOS-2P-stabilized SIV Env trimer accurately reflected the structure of the in situ membrane-bound form of Env trimer on the surface of virions. We determined two in situ structures of SIVmac239 Env trimer on the virion surface by cryo-electron tomography (cryo-ET) and sub-tomogram averaging, one bound to ITS90.0324 and the other ligand-free (Fig. 6, Table 2 and Extended Data Fig. 9). The model determined from the soluble trimer fit well into the cryo-ET densities of ITS90.03-bound and ligand-free reconstructions, with correlation coefficients of 0.71 and 0.62, respectively.
a, A slice of the sub-tomogram average of the ITS90.03-bound SIVmac239 Env on an AT-2-inactivated virion is shown from a view at the apex of the Env looking down the three-fold axis. b, A sub-tomogram average slice is shown, as in a, but rotated 90° to show the side view. c, Cryo-ET volume density, shown from the same view, with each protomer colored as gray, pink and blue. N-linked glycans are colored green, and potential O-linked glycan sites are in red. The neutralizing antibody ITS90.03 is colored purple and tan for the light and heavy chain, respectively. d, As in c, but rotated 90° to a side view to show the angle of approach of ITS90.03. e, The cryo-ET density is shown as a transparent surface with the trimeric atomic model fit. The ITS90.03 antibody is shown from the structure PDB 6TYB after alignment with the trimer by the gp120 core region. f, View of the ITS90.03 crystal structure PDB 6TYB docked into the cryo-ET density. g, A slice of a sub-tomogram average of the SIV ligand-free trimer, displayed in the same orientation as a. h, Cryo-ET volume of the ligand-free trimer as in e. i, The model from the soluble trimer is fit to the density. Extra density is observed for the MPER region, displacing the Env from the membrane by over 10 Å. Little density is observed for residues 512–569.
ITS90.03—one of the few SIV antibodies that has been observed to fully neutralize SIVmac239—bound to the gp120-gp41 interface region, as expected from the previously determined crystal structure of SIVmac239 gp120 core in complex with ITS90.03 and rhCD424, accentuating the three-blade propeller structure of the SIV trimer (Fig. 6a,b). This antibody made minimal contacts with gp41 in the trimer context when aligned by the gp120 chains, although we note that the glycan at N625 must move from its orientation in the soluble structure to accommodate the antibody (Extended Data Fig. 9c–e). The O-glycosylated extended V1 loop could be clearly distinguished in the tomographic density, and density could be observed for all 25 of the N-linked sites of glycosylation found in the soluble structure, although density was noticeably weaker in several sites, including the N160 glycan at the apex (Fig. 6c–f).
The ligand-free in situ structure of the SIV Env trimer was indistinguishable from the in situ SIV trimer bound by ITS90.03, apart from density associated with the antibody (Fig. 6g–i). For both in situ structures, additional density was observed for the region corresponding to membrane-proximal external region (MPER) residues 665–683, defining the MPER to form a pedestal, proximal to the three-fold axis of the trimer and interior to the soluble Env gp41 C-terminal helices, as has been observed for HIV-148 (Extended Data Fig. 9f–k). The MPER pedestal elevated the C terminus of the soluble SIV Env trimer construct (ending at residue 664) over 10 Å from the surface of the virion membrane (Fig. 6i and Extended Data Fig. 9). Altogether, Env features observed on the virion surface, for ligand-free and ITS90.03-bound structures, closely matched those observed in the stabilized soluble SIV-Env trimer.
Discussion
Prefusion-stabilized soluble HIV-1 envelope (Env) trimers22,49,50,51 and their associated analyses, including residue-level structural definition52,53,54,55,56,57, have provided a dramatic leap forward in HIV-1 research from their use as immunogens58,59,60,61,62,63 and as structural templates for mapping antibody epitopes to rationally assess vaccine development64,65,66,67,68,69. Here we obtain a prefusion-stabilized soluble SIVmac Env trimer and determine its structure in complexes with neutralizing antibodies. The revealed SIVmac structure enabled the definition of variable loops and glycan shielding throughout SIV, including details of extended SIV-variable loops and of N- and O-linked glycosylation. We further show the obtained soluble trimer structure to be in the same conformation as both neutralizing antibody-bound and ligand-free in situ structures of Env trimer on SIVmac239 virions.
Understanding SIV, which has infected primates in Africa for thousands of years, provides insight into the long-range evolutionary development of HIV-1. Conservation of variable loops masking and glycan shielding indicates immune evasion to be maintained, despite substantial adaptation, including the evolution of non-pathogenic phenotypes in multiple species70,71,72,73. Env-based immune evasion is thus likely to continue to persist with HIV-1.
The sequence conservation and reduced glycan shielding that we observed for the CD4-binding site throughout SIV provides insight into the impact of receptor function on Env-sequence variation. Most notable, however, may be the sequence conservation of the Env apex close to the trimer three fold and, to a lesser extent, some of the less glycosylated surfaces proximal to the fusion peptide (Extended Data Fig. 6b). The conservation of these regions does not appear to derive from receptor-binding constraints and may instead stem from folding requirements for entry and/or conformational masking. Whatever the cause, the conservation of these sites throughout SIV indicates they are likely to remain conserved in HIV-1, providing additional rationales for targeting these sites with both passive antibody treatment and vaccine development.
Methods
Design of stabilized SIV trimer
We modeled the SIVmac Env structure based on prefusion-closed structures HIV-1 Env, and designed stabilized SIV Env variants, most of which incorporated the SOSIP mutations (for example, cysteine mutations at positions 501 and 605 and a proline mutation at 559) with supplemental disulfides. Additional designs including proline mutations to disrupt secondary structure, as well as cavity-filling mutations and screening of various SIV strains. We screened the designed SIV Env variant mutants for expression and antigenicity in a 96-well transient-expression format74. Mutants that showed improved binding to antibody ITS92.02 by ELISA were selected for purification and further characterization. A design named SOS-2P, which contained SOSIP mutations and a second proline mutation, T569P, displayed the best antigenicity and was selected for structural studies. To enable binding by antibody PGT145, we further altered the SIV Env SOS-2P design to incorporate a K169T mutation.
Protein expression and purification
For immunoglobulin-G (IgG) preparation, DNA sequences of antibody heavy and light chain variable regions were synthesized and subcloned into the pVRC8400 vector, as described previously75. To facilitate the preparation of antigen-binding fragments (Fabs), an HRV3C cleavage site was inserted into the heavy chain hinge region of selected IgG constructs (IgG-3C). IgG expression was carried out by co-transfecting both heavy and light chain plasmids in Expi293F cells (Thermo Fisher) using Turbo293 transfection reagent (Speed BioSystems). On day 6 post transfection, the culture supernatant was collected and loaded onto a protein A column. After washing with phosphate buffered saline (PBS), antibody was eluted with IgG elution buffer (Pierce) and immediately pH-neutralized by addition of a one-tenth volume of 1 M Tris-HCl, pH 8.0. Antibodies used in purification have been published previously, except for ITS92.02, which is unrelated to ITS90 and was isolated from animal IK16, a rhesus macaque vaccinated with a CD4-independent variant of SIVmac23976 that became infected following challenge with SIVsmE660 swarm virus. Briefly, bulk sorted B cells from cryo-preserved peripheral blood mononuclear cells were expanded using B-cell culture conditions optimized for human B cells77. Culture supernatants from expanded B cells were evaluated for neutralization of SIVsmE660.CR54.2A5 (referred to as SIVsmE660.CR54 in the text) Env-pseudotyped virus using the high-throughput NVITAL automated microneutralization assay as previously described78. Immunoglobulin heavy and light chain variable regions from B-cell wells that were positive for SIVsmE660.2A5.CR54.2A5 neutralization were amplified by polymerase chain reaction with reverse transcription (RT-PCR) as previously described79, using rhesus Ig-specific primers80,81.
To generate Fab, IgG-3C was incubated with HRV3C protease at a ratio of 100 units per 10 mg IgG in cleavage buffer containing 150 mM NaCl, 50 mM Tris-HCl, pH 7.5 at 4 °C overnight. After inactivating the HRV3C protease with 1× cOmplete Protease Inhibitor Cocktail (Roche), the digestion mixture was loaded onto a Protein A column (GE Health Science), and Fab from the column flowthrough was collected and concentrated.
Expression plasmids of SIV Env trimers for initial 96-well screening were constructed in a similar manner to that for HIV-1 BG505 Env22 and were expressed in a HEK293S GnTI- cell line as described previously50. To make a complex of SOS-2P Env and ITS92.02 Fab, ITS92.02 IgG-3C was first loaded onto a Protein A column, then supernatant from the expression of SIV Env trimer was flowed through the ITS92.02 IgG-3C-bound protein A column before unbound Env was washed away with PBS. The complex of SOS-2P Env and ITS92.02 Fab was obtained by subsequent HRV3C cleavage (Extended Data Fig. 1e). For the SIV Env complexed with PGT145, SIVmac239 SOS-2P trimer with additional K169T mutation was co-expressed with PGT145 IgG-3C, followed by on-column purification using protein A to bind the PGT145 FC region, then by washing with PBS and HRV3C cleavage.
Preparation of AT2-inactived SIV virions
The 5′ and 3′ half genomes of SIVmac239 were obtained from the HIV Reagent Program, cat. nos. ARP-829 and ARP-830, respectively34. The 3′ half clone contains a premature stop codon in nef and an additional stop codon was added to the 3′ half clone at position 734 of the env gene that results in truncation of the TM protein in virions. The regenerated provirus molecular clone (pSIV239Q734*) was used to produce a transfection-derived infectious stock, which was used to infect the SUPT1-CCR5 cell line (SR5) provided by J. Hoxie (University of Pennsylvania). After infection, the culture required addition of fresh uninfected SR5 cells on three occasions early post infection because of the substantial viral cytopathic effects. A stable, virally infected cell line emerged after about a month in culture and, at 54 days post infection, limiting dilution clones were prepared, and the virus produced by one clone (SIVmac239*/SR5 CL.09) was used in this study. Population-level proviral sequencing of this limiting dilution clone confirmed the retention of the Env and Nef stop codons in addition to two acquired mutations that resulted in amino acid changes: POL:R954G and ENV:G31D relative to the wild-type sequence. The analyzed virus (SIVmac239*/SR5 lot P4465) was treated with 2,2′-dithiodipyridine (Aldrithiol-2, AT-2) to eliminate infectivity while maintaining functional envelope glycoprotein spikes, as described in refs. 82,83. Virus was purified as described in ref. 48.
Virus neutralization assays
Virus neutralization was measured using single round infection of TZM-bl target cells by SIV Env-pseudotyped virus. 293T-derived HIV-1 Env-pseudotyped virus stocks were generated by co-transfection of an Env expression plasmid and a pSG3ΔEnv backbone. Monoclonal antibodies were tested at eight-point fivefold dilutions starting at 50 μg ml−1. Env-pseudotyped virus stocks and antibodies were mixed in a total volume of 50 μl and incubated at 37 °C for 1 h. TZM-bl cells (20 μl, 0.5 million ml−1) were then added to the virus and antibody mixture and incubated at 37 °C for 24 h. Cells were fed with 70 μl cDMEM and incubated for a further 24 h, before being lysed and assessed for luciferase activity (relative light units, RLUs). A nonlinear regression curve was fit using five-parameter hill slope equation. The 50% inhibitory concentration (IC50) was defined as the antibody concentration that caused a 50% reduction in RLU compared to virus control wells after subtraction of background RLU. Measurements were performed in duplicate, and mean values were plotted with error bars indicating s.d.
Negative-stain EM
SIVmac239 SOS-2P trimer–Fab complex samples were diluted in buffer containing 10 mM HEPES, pH 7.0, and 150 mM NaCl to a final trimer concentration of ~0.02 mg ml−1. The sample was adsorbed to freshly glow-discharged carbon-coated grids, rinsed with several drops of the dilution buffer, and stained with 0.75% uranyl formate. Images were recorded with defocus values ranging from −0.3 to −1.5 µm, and EMAN284 and Relion 1.485 were used to obtain reference-free two-dimensional (2D) classification and averaging.
Single-particle cryo-EM
SIVmac239 SOS-2P trimer–Fab complex samples were concentrated to ~1.5 mg ml−1, and 2.3 μl of the complex was deposited on a C-flat grid (protochip.com). The grid was vitrified in ethane with an FEI Vitrobot Mark IV set to a wait time of 30 s, blot time of 3 s and blot force of 1. Automated data collection on a Titan Krios electron microscope was performed with Leginon v3.486 with a Gatan K3 direct detection device. Exposures were collected in movie mode for 2 s, with the total dose of 51.15 e−/Å2 fractionated over 50 raw frames. Images were pre-processed using Appion v3.487,88 during collection; individual frames were aligned and dose-weighted using MotionCor289, and CTFFind490,91 was used to estimate the initial CTF. CryoSPARC 3.392 was used for the final CTF, 2D classifications, ab initio 3D reconstruction, homogeneous refinement and non-uniform 3D refinement. Initial 3D reconstruction and final refinements were performed using C1 symmetry.
Coordinates from PDB 6TYB24 were used for the initial fit to the reconstructed map. This was followed by manual building in Coot v0.993 and simulated annealing and real space refinement in Phenix v1.19.294, and then iterative processing with manual fitting of the coordinates in Coot93. Geometry and map-fitting were evaluated throughout the process using MolProbity95 and EMRinger96. PyMOL v2.4.2 (www.pymol.org) and ChimeraX v1.397 were used to generate structure figures.
Cryo-ET and sub-tomogram averaging
SIVmac239 with delta CT was inactivated with Aldrithiol-2(AT-2). SIVmac239 was incubated with antibody ITS90.03 for 30 min at room temperature. Colloidal gold (6 nm) solution was added to the mixture, and 4 μl of virus sample was placed onto freshly glow-discharged holey carbon grid (Quantifoil Cu R2/1, 200 mesh) for 1 min. The grid was blotted with Whatman filter paper from the back, and was rapidly frozen in liquid ethane using a homemade gravity-driven plunger apparatus.
The frozen-hydrated SIVmac239 samples were imaged at −170 °C on a cryo-electron microscope (Titan Krios, Thermo Fisher Scientific) with a direct detection camera (K3 Summit, Gatan) and an energy filter slit width of 20 eV. The images were captured at a magnification of ×64,000 with a defocus around −4 µm, resulting in 1.38 Å pixel−1 at the specimen level. The tomographic package SerialEM was utilized to collect dose symmetric tilt series at 3° steps from −51° to +51°, with a total dose of ~60 e−/Å2. The dose-fractionated mode was used to generate 8–10 frames per each projection image.
All frames were aligned and motion-corrected using MotionCor289. The drift-corrected tilt series were aligned using 6-nm gold fiducial makers by IMOD98,99. The six binned reconstructions were generated by TOMO3D100. In total, we generated 55 tomograms of SIVmac239 and 105 tomograms of SIVmac239 complex with ITS90.03. To analyze SIVmac239 Env, we first extracted virus particles from the tomograms, then used PyTom template-match101,102 to identify Env spikes. The initial orientation of each Env spike was estimated based on the center of the virus and the center of the Env. To remove junk particles, 3D classification implemented in I3 (0.9.9.3)103,104 was used to generate 100 class averages. After removing junk classes, good class averages were then subjected to the ‘alignment by classification’ method to generate an initial low-resolution structure105. In total, of 60,464 Env spikes in complex with ITS90.03, 32,857 Env spikes were selected for further refinement of unbinned sub-tomograms. Both C3 symmetry and molecular mask were applied to improve the resolution. The resolutions of the maps were estimated by ResMap and the membrane region was masked.
Env-sequence conservation
Sequence conservation was calculated based on the Env sequences of 195 strains from every branch of the primate immunodeficiency virus Env phylogenetic tree, as shown in Fig. 4a. The sequence entropy was calculated for each position in the aligned sequences with the formula Entropy = −∑i p(xi)log(p(xi)), where xi is each of the 20 standard amino acids or gap in the aligned sequences106.
Molecular dynamics simulations and glycan coverage analysis
Molecular dynamics (MD) simulations were carried out for ten glycosylated trimers—with non-sialylated core-1 O-linked glycans on the V1 loop with Man-5 in all potential N-linked glycan sites. The initial systems were prepared by the Glycan Reader & Modeler107 in CHARMM-GUI108. All atom MD simulations were performed using NAMD2.13109, and FreeSASA110 was used to determine the epitope residues of PGT145, the CD4 binding site and V3 region, and their surface area, with 1.4 Å as a probe radius. The program GLYCO111 was used to quantify the glycan shielding of the Env trimers. This program counts the glycan atoms of the protein surface within a 23-Å-distance cutoff from each surface residue, excluding those that are hindered by the protein region. The counted glycan atoms were averaged over 500 ns of MD simulations. The cutoff was determined by analyzing the longest length of the glycans of Man-5 and the disaccharide glycosylated Env trimer, averaged over 500 ns of MD simulations. Epitope-glycan coverage was calculated by the number of glycan atoms of the epitope divided by its surface area. The number of glycan atoms per epitope was counted with the same method as described above. Summation of the SASA of each epitope residue was used as a factor to normalize the number of glycan atoms. Modeling of N-linked glycan shields for other SIV family members was performed using Glycoprotein Builder and manually adjusted through Coot.
Quantification and statistical analysis
Quantification of structural superpositions and alignments was performed with The PyMOL Molecular Graphics System, v2.4.2 (Schrödinger). Buried surface areas were calculated with PISA112. CHARMM-GUI Glycan Reader & Modeler was used for modeling of carbohydrates, and all atom MD simulations were performed using NAMD2.13. Certain graphs were generated using GraphPad Prism v9.0.2.
Material availability
We synthesized DNA for the antibodies and SIV trimer constructs reported in this Article. Any unique mutations are explicitly detailed in the text and methods. Plasmids are available upon requests to P.D.K. (pdkwong@nih.gov) for non-commercial research purposes.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this Article.
Data availability
Cryo-ET maps for the ITS90.03-bound spike and the unliganded spike have been deposited at the Electron Microscopy Data Bank (EMDB) with accession codes EMD-25065 and EMD-25064, respectively. Cryo-EM maps and fitted coordinates have been deposited at the Electron Microscopy Data Bank with accession codes EMD-27718, EMD-27735 and EMD-27631 and to the Protein Data Bank with IDs 8DUA and 8DVD.
Code availability
The program GLYCO is available at https://github.com/myungjinlee/GLYCO.
References
Daniel, M. D. et al. Isolation of T-cell tropic HTLV-III-like retrovirus from macaques. Science 228, 1201–1204 (1985).
Kanki, P. J. et al. Serologic identification and characterization of a macaque T-lymphotropic retrovirus closely related to HTLV-III. Science 228, 1199–1201 (1985).
Aghokeng, A. F. et al. Extensive survey on the prevalence and genetic diversity of SIVs in primate bushmeat provides insights into risks for potential new cross-species transmissions. Infect. Genet. Evol. 10, 386–396 (2010).
Peeters, M., D’Arc, M. & Delaporte, E. Origin and diversity of human retroviruses. AIDS Rev. 16, 23–34 (2014).
Bell, S. M. & Bedford, T. Modern-day SIV viral diversity generated by extensive recombination and cross-species transmission. PLoS Pathog. 13, e1006466 (2017).
Sharp, P. M. & Hahn, B. H. The evolution of HIV-1 and the origin of AIDS. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 2487–2494 (2010).
Worobey, M. et al. Island biogeography reveals the deep history of SIV. Science 329, 1487 (2010).
Compton, A. A., Malik, H. S. & Emerman, M. Host gene evolution traces the evolutionary history of ancient primate lentiviruses. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368, 20120496 (2013).
Gao, F. et al. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397, 436–441 (1999).
Korber, B. et al. Evolutionary and immunological implications of contemporary HIV-1 variation. Br. Med. Bull. 58, 19–42 (2001).
UNAIDS. Global HIV & AIDS Statistics—Fact Sheet (UNAIDS, 2021); https://www.unaids.org/en/resources/fact-sheet
Damond, F. et al. Identification of a highly divergent HIV type 2 and proposal for a change in HIV type 2 classification. AIDS Res. Hum. Retroviruses 20, 666–672 (2004).
Hirsch, V. M., Olmsted, R. A., Murphey-Corb, M., Purcell, R. H. & Johnson, P. R. An African primate lentivirus (SIVsm) closely related to HIV-2. Nature 339, 389–392 (1989).
Harrison, S. C. Viral membrane fusion. Nat. Struct. Mol. Biol. 15, 690–698 (2008).
Wyatt, R. & Sodroski, J. The HIV-1 envelope glycoproteins: fusogens, antigens and immunogens. Science 280, 1884–1888 (1998).
Starcich, B. R. et al. Identification and characterization of conserved and variable regions in the envelope gene of HTLV-III/LAV, the retrovirus of AIDS. Cell 45, 637–648 (1986).
Kwong, P. D. et al. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420, 678–682 (2002).
Wei, X. et al. Antibody neutralization and escape by HIV-1. Nature 422, 307–312 (2003).
Klein, J. S. & Bjorkman, P. J. Few and far between: how HIV may be evading antibody avidity. PLoS Pathog. 6, e1000908 (2010).
Zhu, P. et al. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature 441, 847–852 (2006).
Andrabi, R. et al. The chimpanzee SIV envelope trimer: structure and deployment as an HIV vaccine template. Cell Rep. 27, 2426–2441 e6 (2019).
Sanders, R. W. et al. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 9, e1003618 (2013).
Walker, L. M. et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477, 466–470 (2011).
Gorman, J. et al. Isolation and structure of an antibody that fully neutralizes isolate SIVmac239 reveals functional similarity of SIV and HIV glycan shields. Immunity 51, 724–734 (2019).
Sanders, R. W. et al. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J. Virol. 76, 8875–8889 (2002).
Pallesen, J. et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl Acad. Sci. USA 114, E7348–E7357 (2017).
Wrapp, D. et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367, 1260–1263 (2020).
Yang, Z. N. et al. The crystal structure of the SIV gp41 ectodomain at 1.47-Å resolution. J. Struct. Biol. 126, 131–144 (1999).
Lee, J. H. et al. A broadly neutralizing antibody targets the dynamic HIV envelope trimer apex via a long, rigidified, and anionic β-hairpin structure. Immunity 46, 690–702 (2017).
Liu, Q. et al. Quaternary contact in the initial interaction of CD4 with the HIV-1 envelope trimer. Nat. Struct. Mol. Biol. 24, 370–378 (2017).
McLellan, J. S. et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature 480, 336–343 (2011).
von Bredow, B. et al. Differences in the binding affinity of an HIV-1 V2 apex-specific antibody for the SIVsmm/mac envelope glycoprotein uncouple antibody-dependent cellular cytotoxicity from neutralization. mBio 10, e01255–19 (2019).
Naidu, Y. M. et al. Characterization of infectious molecular clones of simian immunodeficiency virus (SIVmac) and human immunodeficiency virus type 2: persistent infection of rhesus monkeys with molecularly cloned SIVmac. J. Virol. 62, 4691–4696 (1988).
Regier, D. A. & Desrosiers, R. C. The complete nucleotide sequence of a pathogenic molecular clone of simian immunodeficiency virus. AIDS Res. Hum. Retroviruses 6, 1221–1231 (1990).
Roederer, M. et al. Immunological and virological mechanisms of vaccine-mediated protection against SIV and HIV. Nature 505, 502–508 (2014).
Li, H. et al. Envelope residue 375 substitutions in simian-human immunodeficiency viruses enhance CD4 binding and replication in rhesus macaques. Proc. Natl Acad. Sci. USA 113, E3413–E3422 (2016).
O’Brien, S. P. et al. Rational design and in vivo selection of SHIVs encoding transmitted/founder subtype C HIV-1 envelopes. PLoS Pathog. 15, e1007632 (2019).
Steentoft, C. et al. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 32, 1478–1488 (2013).
Silver, Z. A. et al. Discovery of O-linked carbohydrate on HIV-1 envelope and its role in shielding against one category of broadly neutralizing antibodies. Cell Rep. 30, 1862–1869 (2020).
Stansell, E., Canis, K., Haslam, S. M., Dell, A. & Desrosiers, R. C. Simian immunodeficiency virus from the sooty mangabey and rhesus macaque is modified with O-linked carbohydrate. J. Virol. 85, 582–595 (2011).
Stansell, E. et al. Gp120 on HIV-1 virions lacks O-linked carbohydrate. PLoS ONE 10, e0124784 (2015).
Chuang, G. Y. et al. Structural survey of broadly neutralizing antibodies targeting the HIV-1 Env trimer delineates epitope categories and characteristics of recognition. Structure 27, 196–206 (2019).
MacLeod, D. T. et al. Early antibody lineage diversification and independent limb maturation lead to broad HIV-1 neutralization targeting the Env high-mannose patch. Immunity 44, 1215–1226 (2016).
Walker, L. M. et al. A limited number of antibody specificities mediate broad and potent serum neutralization in selected HIV-1 infected individuals. PLoS Pathog. 6, e1001028 (2010).
Arthos, J. et al. HIV-1 envelope protein binds to and signals through integrin α4β7, the gut mucosal homing receptor for peripheral T cells. Nat. Immunol. 9, 301–309 (2008).
Bibollet-Ruche, F. et al. New simian immunodeficiency virus infecting De Brazza’s monkeys (Cercopithecus neglectus): evidence for a cercopithecus monkey virus clade. J. Virol. 78, 7748–7762 (2004).
Zolla-Pazner, S., Alvarez, R., Kong, X. P. & Weiss, S. Vaccine-induced V1V2-specific antibodies control and or protect against infection with HIV, SIV and SHIV. Curr. Opin. HIV AIDS 14, 309–317 (2019).
Li, Z. et al. Subnanometer structures of HIV-1 envelope trimers on aldrithiol-2-inactivated virus particles. Nat. Struct. Mol. Biol. 27, 726–734 (2020).
Torrents de la Pena, A. et al. Improving the immunogenicity of native-like HIV-1 envelope trimers by hyperstabilization. Cell Rep. 20, 1805–1817 (2017).
Kwon, Y. D. et al. Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1 Env. Nat. Struct. Mol. Biol. 22, 522–531 (2015).
Georgiev, I. S. et al. Single-chain soluble BG505.SOSIP gp140 trimers as structural and antigenic mimics of mature closed HIV-1 Env. J. Virol. 89, 5318–5329 (2015).
Julien, J. P. et al. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science 342, 1477–1483 (2013).
Lee, J. H., Ozorowski, G. & Ward, A. B. Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science 351, 1043–1048 (2016).
Lyumkis, D. et al. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science 342, 1484–1490 (2013).
Ozorowski, G. et al. Open and closed structures reveal allostery and pliability in the HIV-1 envelope spike. Nature 547, 360–363 (2017).
Pancera, M. et al. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature 514, 455–461 (2014).
Stewart-Jones, G. B. et al. Trimeric HIV-1-Env structures define glycan shields from clades A, B and G. Cell 165, 813–826 (2016).
Sanders, R. W. et al. HIV-1 VACCINES. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science 349, aac4223 (2015).
de Taeye, S. W. et al. Immunogenicity of stabilized HIV-1 envelope trimers with reduced exposure of non-neutralizing epitopes. Cell 163, 1702–1715 (2015).
Pauthner, M. et al. Elicitation of robust tier 2 neutralizing antibody responses in nonhuman primates by HIV envelope trimer immunization using optimized approaches. Immunity 46, 1073–1088 e6 (2017).
Xu, K. et al. Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1. Nat. Med. 24, 857–867 (2018).
Escolano, A. et al. Immunization expands B cells specific to HIV-1 V3 glycan in mice and macaques. Nature 570, 468–473 (2019).
Steichen, J. M. et al. HIV vaccine design to target germline precursors of glycan-dependent broadly neutralizing antibodies. Immunity 45, 483–496 (2016).
Blattner, C. et al. Structural delineation of a quaternary, cleavage-dependent epitope at the gp41-gp120 interface on intact HIV-1 Env trimers. Immunity 40, 669–680 (2014).
Scharf, L. et al. Antibody 8ANC195 reveals a site of broad vulnerability on the HIV-1 envelope spike. Cell Rep. 7, 785–795 (2014).
Huang, J. et al. Broad and potent HIV-1 neutralization by a human antibody that binds the gp41–gp120 interface. Nature 515, 138–142 (2014).
Julien, J. P. et al. Asymmetric recognition of the HIV-1 trimer by broadly neutralizing antibody PG9. Proc. Natl Acad. Sci. USA 110, 4351–4356 (2013).
Julien, J. P. et al. Broadly neutralizing antibody PGT121 allosterically modulates CD4 binding via recognition of the HIV-1 gp120 V3 base and multiple surrounding glycans. PLoS Pathog. 9, e1003342 (2013).
Kong, L. et al. Complete epitopes for vaccine design derived from a crystal structure of the broadly neutralizing antibodies PGT128 and 8ANC195 in complex with an HIV-1 Env trimer. Acta Crystallogr. D Biol. Crystallogr. 71, 2099–2108 (2015).
Broussard, S. R. et al. Simian immunodeficiency virus replicates to high levels in naturally infected African green monkeys without inducing immunologic or neurologic disease. J. Virol. 75, 2262–2275 (2001).
Chahroudi, A., Bosinger, S. E., Vanderford, T. H., Paiardini, M. & Silvestri, G. Natural SIV hosts: showing AIDS the door. Science 335, 1188–1193 (2012).
Cichutek, K. & Norley, S. Lack of immune suppression in SIV-infected natural hosts. AIDS 7, S25–S35 (1993).
Rey-Cuille, M. A. et al. Simian immunodeficiency virus replicates to high levels in sooty mangabeys without inducing disease. J. Virol. 72, 3872–3886 (1998).
McLellan, J. S. et al. Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science 342, 592–598 (2013).
Wu, X. et al. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science 333, 1593–1602 (2011).
Swanstrom, A. E. et al. Derivation and characterization of a CD4-independent, non-CD4-tropic simian immunodeficiency virus. J. Virol. 90, 4966–4980 (2016).
Huang, J. et al. Isolation of human monoclonal antibodies from peripheral blood B cells. Nat. Protoc. 8, 1907–1915 (2013).
Doria-Rose, N. et al. High throughput HIV-1 microneutralization assay. Protocol Exchange https://doi.org/10.1038/protex.2013.069 (2013).
Longo, N. S. et al. Multiple antibody lineages in one donor target the Glycan-V3 supersite of the HIV-1 envelope glycoprotein and display a preference for quaternary binding. J. Virol. 90, 10574–10586 (2016).
Mason, R. D. et al. Targeted isolation of antibodies directed against major sites of SIV Env vulnerability. PLoS Pathog. 12, e1005537 (2016).
Kong, R. et al. Antibody lineages with vaccine-induced antigen-binding hotspots develop broad HIV neutralization. Cell 178, 567–584 (2019).
Arthur, L. O. et al. Chemical inactivation of retroviral infectivity by targeting nucleocapsid protein zinc fingers: a candidate SIV vaccine. AIDS Res. Hum. Retroviruses 14, S311–S319 (1998).
Rossio, J. L. et al. Inactivation of human immunodeficiency virus type 1 infectivity with preservation of conformational and functional integrity of virion surface proteins. J. Virol. 72, 7992–8001 (1998).
Tang, G. et al. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007).
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Suloway, C. et al. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 151, 41–60 (2005).
Voss, N. R., Yoshioka, C. K., Radermacher, M., Potter, C. S. & Carragher, B. DoG Picker and TiltPicker: software tools to facilitate particle selection in single particle electron microscopy. J. Struct. Biol. 166, 205–213 (2009).
Lander, G. C. et al. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J. Struct. Biol. 166, 95–102 (2009).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Zhang, K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Adams, P. D. et al. Recent developments in the PHENIX software for automated crystallographic structure determination. J. Synchrotron Radiat. 11, 53–55 (2004).
Davis, I. W., Murray, L. W., Richardson, J. S. & Richardson, D. C. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 32, W615–W619 (2004).
Barad, B. A. et al. EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy. Nat. Methods 12, 943–946 (2015).
Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators and developers. Protein Sci. 30, 70–82 (2021).
Mastronarde, D. N. & Held, S. R. Automated tilt series alignment and tomographic reconstruction in IMOD. J. Struct. Biol. 197, 102–113 (2017).
Mastronarde, D. N. Correction for non-perpendicularity of beam and tilt axis in tomographic reconstructions with the IMOD package. J. Microsc. 230, 212–217 (2008).
Agulleiro, J. I. & Fernandez, J. J. Tomo3D 2.0–exploitation of advanced vector extensions (AVX) for 3D reconstruction. J. Struct. Biol. 189, 147–152 (2015).
Hrabe, T. Localize.pytom: a modern webserver for cryo-electron tomography. Nucleic Acids Res. 43, W231–W236 (2015).
Hrabe, T. et al. PyTom: a python-based toolbox for localization of macromolecules in cryo-electron tomograms and subtomogram analysis. J. Struct. Biol. 178, 177–188 (2012).
Winkler, H. 3D reconstruction and processing of volumetric data in cryo-electron tomography. J. Struct. Biol. 157, 126–137 (2007).
Winkler, H. et al. Tomographic subvolume alignment and subvolume classification applied to myosin V and SIV envelope spikes. J. Struct. Biol. 165, 64–77 (2009).
Liu, J., Wright, E. R. & Winkler, H. 3D visualization of HIV virions by cryoelectron tomography. Methods Enzymol. 483, 267–290 (2010).
Xu, J. et al. Nanobodies from camelid mice and llamas neutralize SARS-CoV-2 variants. Nature 595, 278–282 (2021).
Park, S. J. et al. CHARMM-GUI Glycan Modeler for modeling and simulation of carbohydrates and glycoconjugates. Glycobiology 29, 320–331 (2019).
Jo, S., Kim, T., Iyer, V. G. & Im, W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 (2008).
Phillips, J. C. et al. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005).
Mitternacht, S. FreeSASA: an open source C library for solvent accessible surface area calculations. F1000Res 5, 189 (2016).
Lee, M., Reveiz, M., Rawi, R., Kwong, P. D. & Chuang, G. Y. GLYCO: a tool to quantify glycan shielding of glycosylated proteins. Bioinformatics 38, 1152–1154 (2021).
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007).
Acknowledgements
We thank J. Hoxie of the University of Pennsylvania for the SUPT1-CCR5 cell line, J. Stuckey of the Vaccine Research Center for assistance with figures, members of the Structural Biology Section and Structural Bioinformatics Core, Vaccine Research Center for discussions or comments on the manuscript, and the HIV Reagent Program for ARP-829 and ARP-830. Support for this work was provided by the Intramural Research Programs of the Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health and from the National Cancer Institute, National Institutes of Health under contract nos. HHSN261200800001E and 75N91019D00024. Some of this work was performed at the Simons Electron Microscopy Center and the National Resource for Automated Molecular Microscopy, located at the New York Structural Biology Center, supported by grants from the Simons Foundation (SF349247) and NIH National Institute of General Medical Sciences (GM103310), with additional support from NYSTAR and the New York State Assembly. Cryo-ET work was performed at the Yale Cryo-EM Resource, which is funded in part by NIH grants nos. 1S10OD023603-01A1 and R01AI150560. This work utilized the computational resources of the NIH HPC Biowulf cluster.
Author information
Authors and Affiliations
Contributions
Conceptualization was provided by J.G., T.Z., R.D.M., M.R. and P.D.K., ITS92.02 isolation and neutralization by R.D.M. and H.C.W., protein expression by Y.Y., B.Z. and R.V., protein purification by T.Z. and A.F.N., cryo-EM structure determination and analysis by J.G., molecular dynamics by M.L., negative stain electron microscopy by Y.T., SIV Env divergence by T.B., virion production and characterization by J.W.B., B.F.K. and J.D.L. and cryo-ET structure determination and analysis by C.W. and J.L. The first draft was written by J.G., and all authors provided feedback and edits.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Structural & Molecular Biology thanks James Hoxie and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Beth Moorefield, in collaboration with the Nature Structural & Molecular Biology team. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Antigenic screening and HRV 3 C purification through capture of co-expression enriches for minority population of trimeric Env.
a, Sequence homology between HIV-1 and several SIV strains is shown. b, The 96-well screening results are presented as red-white heatmap with the top scoring constructs enlarged on the right panel showing their specific values for HIV-1 and SIV antibodies. The bottom-right table summarized the screening antibody epitopes. The ITS92 epitope was unknown during the screening and the accompanying structures revealed it to be a gp41 conformationally dependent epitope. c, Negative stain 2D images show a small population of trimers, highlighted here in the green squares. d, Schematic of the purification strategy is shown with on column binding of the supernatant to ITS92.02 followed by HRV 3 C cleavage, allowing the avoidance of harsh purification buffers. A similar strategy was used for PGT145 but included co-expression with the HRV 3 C IgG.
Extended Data Fig. 2 Cryo-EM analysis of SIVE660.CR54 SOS-2P in complex with ITS92.02 reveals a trimer with high flexibility at the apex.
a, Representative micrograph and CTF of the micrograph are shown. 8,259 micrographs were collected in total. b, Representative 2D class averages are shown. c, The gold-standard Fourier shell correlation resulted in a resolution of 4.32 Å using non-uniform refinement with C3 symmetry. d, The orientations of all particles used in the final refinement are shown as a heatmap. e, The local resolution of the full map is shown generated through cryoSPARC using an FSC cutoff of 0.5. f, A wild-type sequence alignment near the ITS92.02 epitope is shown highlighting residues within a 5 Å footprint in red. We note that non-conserved residue E613 forms a salt bridge with R94 of the ITS92.02 heavy chain. g, The prefusion (magenta) and postfusion (gray, PDB ID 1QBZ) conformations of gp41 protomers are aligned with the prefusion footprint residues colored red and the corresponding residues in the postfusion conformation are shown in blue. Footprint residues between positions 638 and 652 (HXB2 numbering) align well, residues from 608–614 extend away from the helical region of the epitope.
Extended Data Fig. 3 Cryo-EM analysis of SIVmac239 SOS-2P in complex with PGT145 contains a stabilized apex region.
a, Representative micrograph and CTF of the micrograph are shown. 6,255 micrographs total were collected. b, Representative 2D class averages are shown. c, The gold-standard Fourier shell correlation resulted in a resolution of 4.12 Å using non-uniform refinement with C1 symmetry. d, The orientations of all particles used in the final refinement are shown as a heatmap. e, The local resolution of the full map is shown in two contours. Maps were generated through cryoSPARC using an FSC cutoff of 0.5.
Extended Data Fig. 4 SIVmac239 sequence and structural comparisons.
a, The SIVmac239 trimer aligns well to the CD4-bound SIVmac239 core (PDB: ID 6TYB). The glycan shield shows strong similarity for most glycans available in the core with notable differences at positions 47 and 464 where sequons were not glycosylated in the core and position 88 where the gp41 glycan at position N625 overlaps in space with N88 of the core. b, A sequence alignment of HIV-1 (BG505) and SIV (mac239) are shown with secondary structure shown below. Notable positions are indicated for SOSIP mutations. c, The Cα residue-by-residue distances are shown for HIV-1 and SIVcpz compared to residues of SIVmac239 indicating areas of close structural alignment and highlighting regions that diverge significantly.
Extended Data Fig. 5 CD4 binding details in SIVmac239 and HIV-1.
a, (left) SIVmac239 gp120 from the trimer structure (magenta) is shown aligned to the CD4-bound SIVmac239 gp120 core (gray). (right) SIVmac239 gp120 from the trimer structure (magenta) is shown aligned to an HIV-1 BG505 gp120 from a trimer (light blue). b, (left) The CD4 (yellow) binding site is shown corresponding to the structural alignments in panel a. W427 adjusts out of the pocket that the F43 of CD4 inserts into. (right) Residue 375, used to confer rhCD4 binding to SHIV, is shown in red. Relative positions of W427 in SIV and HIV-1 are shown proximal to the F43 pocket. c, Conformations of the region from residues 50–80 are shown. The region adopts various conformations in SIV and HIV-1 and undergoes a switch upon CD4 binding.
Extended Data Fig. 6 Details of the primate immunodeficiency viruses conserved features.
a, An additional disulfide was observed in one branch which placed cys at residues 165 and 168, stabilizing the turn between the B and C strands of V1V2. b, Disulfides observed in the hypervariable V1 loop are highlighted in purple. Due to poor alignment confidence in this region the sequences are shown from residue cys157 conserved in all viruses in the tree. c, The HIV-2 glycan shield is modeled as in Fig. 5e with minor deviations from SIVmac239. d, The fusion peptide region of SIVmac239 adopts a helical structure and abuts the core of the Env contrary to the flexible conformations observed in HIV-1 Env (BG505, PDB ID: 5FYL). We note that the FP is one residue longer in the viruses more proximal to HIV-1.
Extended Data Fig. 7 Impact of O-linked saccharides on glycan shielding.
a, Glycan shielding overlaid on SIVmac239 surfaces viewing from side (top panel) and top (bottom panel). b, Glycan coverage of epitope CD4bs (CD4 binding site), PGT145 and V3 regions of SIVmac239. The epitope regions are shown (top middle and top right panels) with the same color code as the plot (top left).
Extended Data Fig. 8 N-linked glycan conservation across diverse primate immunodeficiency viruses.
(top) Bar graph depicting the percent conservation of N-linked glycan sequon positions (HXB2 numbering from 1–683) for the 34 viruses depicted in the phylogenetic tree of Fig. 4. Hypervariable regions extending beyond HXB2 numbering are excluded due to high variability and low confidence alignments. * at positions 143 and 185 denotes artificially high bars due to multiple glycans in inserts. Dotted horizontal lines are shown at 50% and 75% conservation and locations above these thresholds are labeled. (bottom) A detailed plot of sequon locations for all 34 sequences in the phylogenetic tree of Fig. 4. Locations are colored as in Fig. 4b.
Extended Data Fig. 9 Cryo-ET density fit to the model with MPER region extended from the membrane.
a, A slice through a representative tomogram shows multiple SIV virions with Env spikes, black scale bar in the bottom right represents 50 nm. 55 tomograms of SIVmac239 and 105 tomograms of SIVmac239 complex with ITS90.03 were collected. b, FSC curves of the ITS90.03-bound (blue) and unliganded SIV (black) sub-tomogram averages. c, Superposition of the ITS90.03 bound crystal structure with that of the SIV trimer based on the gp120 alignment. d, Glycan N625 of the SIVmac239 trimer must re-orient to accommodate the binding of ITS90.03. e, The V1 insertion region is observed in the ITS90.03-bound and unliganded cryo-ET density. f, Side view of the gp41 density with the gp41 built from the soluble map fit to the density. g, Side view to panel d is shown. h, The pedestal at the base of the gp41 region in HIV-1 Bal is shown in yellow, segmented from density of EMDB map EMD-21412. Coordinates are shown for BG505 SOSIP.664. i, The side of gp41 is shown with the MPER region density shown in gold. j, The fit of the MPER region in relation to the membrane surface density. k, The pedestal at the base of the gp41 region in HIV is shown in yellow with membrane density shown.
Supplementary information
Rights and permissions
About this article
Cite this article
Gorman, J., Wang, C., Mason, R.D. et al. Cryo-EM structures of prefusion SIV envelope trimer. Nat Struct Mol Biol 29, 1080–1091 (2022). https://doi.org/10.1038/s41594-022-00852-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-022-00852-1