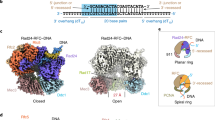
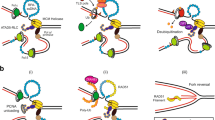
Abstract
Single-stranded or double-stranded DNA junctions with recessed 5′ ends serve as loading sites for the checkpoint clamp, 9-1-1, which mediates activation of the apical checkpoint kinase, ATRMec1. However, the basis for 9-1-1’s recruitment to 5′ junctions is unclear. Here, we present structures of the yeast checkpoint clamp loader, Rad24-replication factor C (RFC), in complex with 9-1-1 and a 5′ junction and in a post-ATP-hydrolysis state. Unexpectedly, 9-1-1 adopts both closed and planar open states in the presence of Rad24-RFC and DNA. Moreover, Rad24-RFC associates with the DNA junction in the opposite orientation of processivity clamp loaders with Rad24 exclusively coordinating the double-stranded region. ATP hydrolysis stimulates conformational changes in Rad24-RFC, leading to disengagement of DNA-loaded 9-1-1. Together, these structures explain 9-1-1’s recruitment to 5′ junctions and reveal new principles of sliding clamp loading.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Cryo-EM maps and atomic coordinates have been deposited with the Electron Microscopy Data Bank and PDB under accession codes EMD-25422 and PDB 7ST9 for Rad24-RFC–9-1-1 in the open state, code EMD-25424 for the Mec3 focus refinement of Rad24-RFC–9-1-1 in the open state, codes EMD-25423 and PDB 7STB for Rad24-RFC–9-1-1 in the closed state, codes EMDB-25426 and PDB 7STE for Rad24-RFCADP, and code EMD-25425 for the Rad24 AAA+ focus refinement of Rad24-RFCADP. Datasets from PDB used in this study include 1SXJ and 3A1J. Source data are provided with this paper.
References
Ciccia, A. & Elledge, S. J. The DNA damage response: making it safe to play with knives. Mol. Cell 40, 179–204 (2010).
Pardo, B., Crabbe, L. & Pasero, P. Signaling pathways of replication stress in yeast. FEMS Yeast Res. 17, https://doi.org/10.1093/femsyr/fow101 (2017).
Saldivar, J. C., Cortez, D. & Cimprich, K. A. The essential kinase ATR: ensuring faithful duplication of a challenging genome. Nat. Rev. Mol. Cell Biol. 18, 622–636 (2017).
Ellison, V. & Stillman, B. Biochemical characterization of DNA damage checkpoint complexes: clamp loader and clamp complexes with specificity for 5′ recessed DNA. PLoS Biol. 1, E33 (2003).
Majka, J., Binz, S. K., Wold, M. S. & Burgers, P. M. Replication protein A directs loading of the DNA damage checkpoint clamp to 5′-DNA junctions. J. Biol. Chem. 281, 27855–27861 (2006).
Zou, L. Four pillars of the S-phase checkpoint. Genes Dev. 27, 227–233 (2013).
Gobbini, E., Casari, E., Colombo, C. V., Bonetti, D. & Longhese, M. P. The 9-1-1 complex controls Mre11 nuclease and checkpoint activation during short-range resection of DNA double-strand breaks. Cell Rep. 33, 108287 (2020).
Ngo, G. H. P., Balakrishnan, L., Dubarry, M., Campbell, J. L. & Lydall, D. The 9-1-1 checkpoint clamp stimulates DNA resection by Dna2-Sgs1 and Exo1. Nucleic Acids Res. 42, 10516–10528 (2014).
van Schendel, R., Romeijn, R., Buijs, H. & Tijsterman, M. Preservation of lagging strand integrity at sites of stalled replication by Pol α-primase and 9-1-1 complex. Sci. Adv. 7, eabf2278 (2021).
Dore, A. S., Kilkenny, M. L., Rzechorzek, N. J. & Pearl, L. H. Crystal structure of the rad9-rad1-hus1 DNA damage checkpoint complex–implications for clamp loading and regulation. Mol. Cell 34, 735–745 (2009).
Sohn, S. Y. & Cho, Y. Crystal structure of the human rad9-hus1-rad1 clamp. J. Mol. Biol. 390, 490–502 (2009).
Xu, M. et al. Structure and functional implications of the human Rad9-Hus1-Rad1 cell cycle checkpoint complex. J. Biol. Chem. 284, 20457–20461 (2009).
Bermudez, V. P. et al. Loading of the human 9-1-1 checkpoint complex onto DNA by the checkpoint clamp loader hRad17-replication factor C complex in vitro. Proc. Natl Acad. Sci. USA 100, 1633–1638 (2003).
Majka, J. & Burgers, P. M. Yeast Rad17/Mec3/Ddc1: a sliding clamp for the DNA damage checkpoint. Proc. Natl Acad. Sci. USA 100, 2249–2254 (2003).
Lee, K. Y. & Park, S. H. Eukaryotic clamp loaders and unloaders in the maintenance of genome stability. Exp. Mol. Med. 52, 1948–1958 (2020).
Kelch, B. A. Review: The lord of the rings: structure and mechanism of the sliding clamp loader. Biopolymers 105, 532–546 (2016).
Bowman, G. D., O’Donnell, M. & Kuriyan, J. Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex. Nature 429, 724–730 (2004).
Gaubitz, C. et al. Structure of the human clamp loader reveals an autoinhibited conformation of a substrate-bound AAA+ switch. Proc. Natl Acad. Sci. USA 117, 23571–23580 (2020).
Devbhandari, S., Jiang, J., Kumar, C., Whitehouse, I. & Remus, D. Chromatin constrains the initiation and elongation of DNA replication. Mol. Cell 65, 131–141 (2017).
Kelch, B. A., Makino, D. L., O’Donnell, M. & Kuriyan, J. How a DNA polymerase clamp loader opens a sliding clamp. Science 334, 1675–1680 (2011).
Michael, W. M., Ott, R., Fanning, E. & Newport, J. Activation of the DNA replication checkpoint through RNA synthesis by primase. Science 289, 2133–2137 (2000).
MacDougall, C. A., Byun, T. S., Van, C., Yee, M. C. & Cimprich, K. A. The structural determinants of checkpoint activation. Genes Dev. 21, 898–903 (2007).
Van, C., Yan, S., Michael, W. M., Waga, S. & Cimprich, K. A. Continued primer synthesis at stalled replication forks contributes to checkpoint activation. J. Cell Biol. 189, 233–246 (2010).
Baretic, D. et al. Cryo-EM structure of the fork protection complex bound to CMG at a replication fork. Mol. Cell 78, 926–940 e913 (2020).
Gao, Y. et al. Structures and operating principles of the replisome. Science https://doi.org/10.1126/science.aav7003 (2019).
Lee, J. Y. & Yang, W. UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell 127, 1349–1360 (2006).
Majka, J., Chung, B. Y. & Burgers, P. M. Requirement for ATP by the DNA damage checkpoint clamp loader. J. Biol. Chem. 279, 20921–20926 (2004).
Naiki, T., Shimomura, T., Kondo, T., Matsumoto, K. & Sugimoto, K. Rfc5, in cooperation with rad24, controls DNA damage checkpoints throughout the cell cycle in Saccharomyces cerevisiae. Mol. Cell. Biol. 20, 5888–5896 (2000).
Majka, J., Niedziela-Majka, A. & Burgers, P. M. The checkpoint clamp activates Mec1 kinase during initiation of the DNA damage checkpoint. Mol. Cell 24, 891–901 (2006).
Kazmirski, S. L., Zhao, Y., Bowman, G. D., O’Donnell, M. & Kuriyan, J. Out-of-plane motions in open sliding clamps: molecular dynamics simulations of eukaryotic and archaeal proliferating cell nuclear antigen. Proc. Natl Acad. Sci. USA 102, 13801–13806 (2005).
Miyata, T. et al. Open clamp structure in the clamp-loading complex visualized by electron microscopic image analysis. Proc. Natl Acad. Sci. USA 102, 13795–13800 (2005).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Scheres, S. H. Processing of structurally heterogeneous cryo-EM sata in RELION. Methods Enzymol. 579, 125–157 (2016).
Punjani, A., Zhang, H. & Fleet, D. J. Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods 17, 1214–1221 (2020).
Terwilliger, T. C., Ludtke, S. J., Read, R. J., Adams, P. D. & Afonine, P. V. Improvement of cryo-EM maps by density modification. Nat. Methods 17, 923–927 (2020).
Pettersen, E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 66, 486–501 (2010).
Croll, T. I. ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr. D. Struct. Biol. 74, 519–530 (2018).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D. Struct. Biol. 75, 861–877 (2019).
Goddard, T. D. et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018).
Acknowledgements
We thank M. de la Cruz at the Memorial Sloan Kettering Cancer Center (MSKCC) Richard Rifkind Center for cryo-EM for assistance with data collection and the MSKCC HPC group for assistance with data processing. This work was supported by NIH-NCI Cancer Center Support grant nos. P30 CA008748 (D.R. and R.K.H.), NIGMS R01-GM107239 (D.R.) and NIGMS R01-GM127428 (D.R.), and the Josie Robertson Investigators Program (R.K.H.). M.S. is a Walter Benjamin Fellow of the Deutsche Forschungsgemeinschaft.
Author information
Authors and Affiliations
Contributions
J.C.C., M.S., D.R. and R.K.H. conceptualized, performed and analyzed the experiments. J.C.C. purified all proteins. M.S. collected and processed cryo-EM data. M.S and R.K.H. built models. All authors contributed to writing the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Structural and Molecular Biology thanks David Jeruzalmi and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Beth Moorefield was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Purification and analysis of Rad24-RFC:9-1-1.
a, Representative Coomassie-stained SDS-PAGE analysis of purified S. cerevisiae Rad24-RFC, 9-1-1 and RPA. The * corresponds to truncated form of Rad24. b, ATPase activity of Rad24-RFC in the presence of 9-1-1 and DNA (black circles), Rad24-RFC in the presence of DNA (teal triangles), Rad24-RFC in the presence of 9-1-1 (purple triangles) and 9-1-1 and DNA (pink squares). Experiments are shown in triplicate. c, Schematic for the assembly of Rad24-RFC:9-1-1:DNA in the presence of ATPγS. d, Representative silver-stained SDS-PAGE analysis of Rad24-RFC:9-1-1 fractions following glycerol gradient (10-35%) centrifugation. Fractions pooled for Cryo-EM analysis are denoted with *. e, Representative Coomassie-stained SDS-PAGE analysis of purified S. cerevisiae Rad24-RFC:9-1-1:DNA. Fractionation experiments shown in panels d and e have been repeated at least three times.
Extended Data Fig. 2 Validation of Rad24-RFC:9-1-1 structures.
a, Representative cryo-EM image of vitrified Rad24-RFC:9-1-1:DNA. b, Representative two-dimensional averages of Rad24-RFC:9-1-1. c, Plot of Fourier shell correlations between two independent open state half-maps (blue), two independent closed state half-maps (red), two independent Mec3 focus half-maps (brown), the open state map and open state atomic model (cyan), and the closed state map and closed state atomic model (magenta). d-f Cryo-EM density maps of open (d), Mec3-focused refined (e) and closed (f) maps colored by local resolution in Å. Region included in the mask for the Mec3 focused refinement (e) is denoted by the dashed line in d.
Extended Data Fig. 3 Comparison of Rad24-RFC:9-1-1 with RFC-PCNA.
a-b, Structures of Rad24-RFC:9-1-1 (a) and RFC-PCNA (b, PDB:1SXJ) colored by subunit with Rad24-RFC / RFC in surface depiction and 9-1-1 / PCNA in ribbon depiction. The channel formed between the AAA + , collar and A’ domains of Rad24 in a is highlighted by the dashed oval. c, Superposition of Rad24-RFC (colored by domain) with RFC (gray). The models are aligned by the collar domains. d, Superposition of Rad24-RFC subunits (colored by subunit) with RFC subunits (grey). The subunits are aligned by their collar domains. For Rad24 and Rfc1, the A’ domains are removed for clarity. e, Nucleotide-binding sites in the AAA + domains Rad24-RFC:9-1-1. Nucleotides and residues whose side chains coordinate the nucleotide are shown in sticks.
Extended Data Fig. 4 Coordination of DNA by Rad24-RFC:9-1-1.
a, Rad24-RFC:9-1-1 surface colored by electrostatic surface potential. DNA is shown as cartoon and Rad24 channel is highlighted by the dashed oval. b, Interactions between ssDNA backbone and conserved arginine residues in Rfc2-5 establish the spiral B-form like conformation of the ssDNA. Arginine side chains are shown as spheres and Rad24 is removed for clarity. c, Superposition of Rad24 in open Rad24-RFC:9-1-1 (colored by subunit) with Rfc1 in RFC-PCNA (colored in grey, PDB:1SXJ), aligned by the AAA + domains. Single-stranded DNA is shown as cartoon. Dashed oval highlights insertion in the AAA + domain of Rad24 that occludes double-stranded DNA from occupying the central cavity of Rad24-RFC. Rfc2-5 are removed for clarity. d, Alignment of residues that coordinate the 5’ junction in S. cerevisiae Rad24 with Rad17 from X. laevis, M. musculus, and H. sapiens. Arrows denote conserved Phe340, which serves as the pin, and conserved Lys345, which coordinates the 5’ phosphate. e, Alignment of residues that coordinate the single-stranded DNA in S. cerevisiae Rfc2, Rfc3 and Rfc4 with RFC4, RFC5 and RFC2 from H. sapiens. Arrows denote conserved isoleucine and arginine residues that coordinate the DNA backbone.
Extended Data Fig. 5 Comparison of open and closed states of Rad24-RFC:9-1-1.
a, Superposition of open (colored by subunit) and closed states of Rad24-RFC:9-1-1 (colored in grey), shown as two views. b, Structure of 9-1-1 in the open state of Rad24-RFC:9-1-1, colored by RMSD in Å. RMSD is calculated on per-subunit basis between the open and closed states of Rad24-RFC:9-1-1. c-d Superposition of 9-1-1 clamp in Rad24-RFC:9-1-1 in closed (c) and open (d) states (colored by subunit) with a structure of human 9-1-1 clamp (colored in grey, PDB:3A1J).
Extended Data Fig. 6 Validation of Rad24-RFCADP structure.
a, Representative cryo-EM image of vitrified Rad24-RFC in the presence of ADP. b, Representative two-dimensional averages of Rad24-RFCADP. c, Plot of Fourier shell correlations between two independent consensus half-maps (blue), two independent Rad24 focus refinement half-maps (brown), and the consensus map and model (cyan). d-e, Cryo-EM density maps of consensus (d) and Rad24 focus refinement (e) maps colored by local resolution in Å. Region included in the mask for the Rad24 AAA + focused refinement (e) is denoted by the dashed line in d.
Extended Data Fig. 7 Conformational changes induced by ATP hydrolysis in Rad24-RFC.
a, Superposition of Rad24-RFC in Rad24-RFCADP (colored by subunit) and Rad24-RFC:9-1-1 (colored in grey) shown in two views. b, Nucleotide-binding sites in the AAA + domains of Rad24-RFCADP. Nucleotides are shown as sticks. c, Superposition of the AAA + inter-domain domain interfaces in Rad24-RFCADP (colored by subunit) and Rad24-RFC:9-1-1 (colored in grey). Nucleotides are shown as sticks.
Supplementary information
Supplementary Information
Supplementary Tables 1–3.
Supplementary Video 1
Morph between open and closed states of Rad24-RFC–9-1-1.
Supplementary Video 2
Morph of 9-1-1 in the closed and open states of Rad24-RFC–9-1-1.
Supplementary Video 3
Morph of Rad24-RFC between ATP-bound state in Rad24-RFC–9-1-1 and ADP-bound state in Rad24-RFCADP.
Source data
Source Data Extended Data Fig. 1
Source data for ATPase assay.
Source Data Extended Data Fig. 1
Unprocessed SDS–PAGE.
Rights and permissions
About this article
Cite this article
Castaneda, J.C., Schrecker, M., Remus, D. et al. Mechanisms of loading and release of the 9-1-1 checkpoint clamp. Nat Struct Mol Biol 29, 369–375 (2022). https://doi.org/10.1038/s41594-022-00741-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-022-00741-7
This article is cited by
-
The partner-swapping sliding clamp loader exposed
Nature Structural & Molecular Biology (2022)