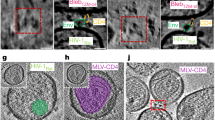
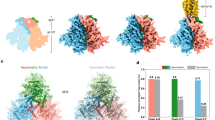
Abstract
The HIV-1 envelope glycoprotein (Env) trimer, composed of gp120 and gp41 subunits, mediates viral entry into cells. Recombinant Env trimers have been studied structurally, but characterization of Env embedded in intact virus membranes has been limited to low resolution. Here, we deploy cryo-electron tomography and subtomogram averaging to determine the structures of Env trimers on aldrithiol-2 (AT-2)-inactivated virions in ligand-free, antibody-bound and CD4-bound forms at subnanometer resolution. Tomographic reconstructions document molecular features consistent with high-resolution structures of engineered soluble and detergent-solubilized Env trimers. One of three conformational states previously predicted by smFRET was not observed by cryo-ET, potentially owing to AT-2 inactivation. We did observe Env trimers to open in situ in response to CD4 binding, with an outward movement of gp120-variable loops and an extension of a critical gp41 helix. Overall features of Env trimer embedded in AT-2-treated virions appear well-represented by current engineered trimers.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Harrison, S. C. Viral membrane fusion. Virology 479–480, 498–507 (2015).
Blumenthal, R., Durell, S. & Viard, M. HIV entry and envelope glycoprotein-mediated fusion. J. Biol. Chem. 287, 40841–40849 (2012).
Ozorowski, G. et al. Open and closed structures reveal allostery and pliability in the HIV-1 envelope spike. Nature 547, 360–363 (2017).
Kwong, P. D. et al. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393, 648–659 (1998).
Rizzuto, C. D. et al. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science 280, 1949–1953 (1998).
Shaik, M. M. et al. Structural basis of coreceptor recognition by HIV-1 envelope spike. Nature 565, 318–323 (2019).
Chan, D. C., Fass, D., Berger, J. M. & Kim, P. S. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89, 263–273 (1997).
Weissenhorn, W., Dessen, A., Harrison, S. C., Skehel, J. J. & Wiley, D. C. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387, 426–430 (1997).
Kwon, Y. D. et al. Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1 Env. Nat. Struct. Mol. Biol. 22, 522–531 (2015).
Julien, J. P. et al. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science 342, 1477–1483 (2013).
Lyumkis, D. et al. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science 342, 1484–1490 (2013).
Pancera, M. et al. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature 514, 455–461 (2014).
Torrents de la Peña, A. et al. Improving the immunogenicity of native-like HIV-1 envelope trimers by hyperstabilization. Cell Rep. 20, 1805–1817 (2017).
Wang, H. et al. Cryo-EM structure of a CD4-bound open HIV-1 envelope trimer reveals structural rearrangements of the gp120 V1V2 loop. Proc. Natl Acad. Sci. USA 113, E7151–E7158 (2016).
Wang, H., Barnes, C. O., Yang, Z., Nussenzweig, M. C. & Bjorkman, P. J. Partially open HIV-1 envelope structures exhibit conformational changes relevant for coreceptor binding and fusion. Cell Host Microbe 24, 579–592.e4 (2018).
Yang, Z., Wang, H., Liu, A. Z., Gristick, H. B. & Bjorkman, P. J. Asymmetric opening of HIV-1 Env bound to CD4 and a coreceptor-mimicking antibody. Nat. Struct. Mol. Biol. 26, 1167–1175 (2019).
Binley, J. M. et al. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J. Virol. 74, 627–643 (2000).
Sanders, R. W. et al. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J. Virol. 76, 8875–8889 (2002).
Sanders, R. W. et al. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 9, e1003618 (2013).
Chuang, G. Y. et al. Structure-based design of a soluble prefusion-closed HIV-1 Env trimer with reduced CD4 affinity and improved immunogenicity. J. Virol. 91, e02268-16 (2017).
Lai, Y. T. et al. Lattice engineering enables definition of molecular features allowing for potent small-molecule inhibition of HIV-1 entry. Nat. Commun. 10, 47 (2019).
Rutten, L. et al. A universal approach to optimize the folding and stability of prefusion-closed HIV-1 envelope trimers. Cell Rep. 23, 584–595 (2018).
Sliepen, K. et al. Structure and immunogenicity of a stabilized HIV-1 envelope trimer based on a group-M consensus sequence. Nat. Commun. 10, 2355 (2019).
Guenaga, J. et al. Glycine substitution at helix-to-coil transitions facilitates the structural determination of a stabilized subtype C HIV envelope glycoprotein. Immunity 46, 792–803.e3 (2017).
Sarkar, A. et al. Structure of a cleavage-independent HIV Env recapitulates the glycoprotein architecture of the native cleaved trimer. Nat. Commun. 9, 1956 (2018).
Kong, L. et al. Uncleaved prefusion-optimized gp140 trimers derived from analysis of HIV-1 envelope metastability. Nat. Commun. 7, 12040 (2016).
Lee, J. H., Ozorowski, G. & Ward, A. B. Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science 351, 1043–1048 (2016).
Pan, J., Peng, H., Chen, B. & Harrison, S. C. Cryo-EM structure of full-length HIV-1 Env bound with the Fab of antibody PG16. J. Mol. Biol. 14, 1158–1168 (2020).
Lee, J. H. et al. A broadly neutralizing antibody targets the dynamic HIV envelope trimer apex via a long, rigidified, and anionic β-hairpin structure. Immunity 46, 690–702 (2017).
Huang, C. C. et al. Structure of a V3-containing HIV-1 gp120 core. Science 310, 1025–8 (2005).
Liu, Q. et al. Quaternary contact in the initial interaction of CD4 with the HIV-1 envelope trimer. Nat. Struct. Mol. Biol. 24, 370–378 (2017).
Ma, X. et al. HIV-1 Env trimer opens through an asymmetric intermediate in which individual protomers adopt distinct conformations. Elife 7, e34271 (2018).
Khasnis, M. D., Halkidis, K., Bhardwaj, A. & Root, M. J. Receptor activation of HIV-1 Env leads to asymmetric exposure of the gp41 trimer. PLoS Pathog. 12, e1006098 (2016).
Zhu, P. et al. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature 441, 847–852 (2006).
Liu, J., Bartesaghi, A., Borgnia, M. J., Sapiro, G. & Subramaniam, S. Molecular architecture of native HIV-1 gp120 trimers. Nature 455, 109–113 (2008).
Zanetti, G., Briggs, J. A. G., Grünewald, K., Sattentau, Q. J. & Fuller, S. D. Cryo-electron tomographic structure of an immunodeficiency virus envelope complex in situ. PLoS Pathog. 2, e83 (2006).
Lu, M. et al. Associating HIV-1 envelope glycoprotein structures with states on the virus observed by smFRET. Nature 568, 415–419 (2019).
Munro, J. B. et al. Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science 346, 759–763 (2014).
Rossio, J. L. et al. Inactivation of human immunodeficiency virus type 1 infectivity with preservation of conformational and functional integrity of virion surface proteins. J. Virol. 72, 7992–8001 (1998).
Chertova, E. et al. Envelope glycoprotein incorporation, not shedding of surface envelope glycoprotein (gp120/SU), is the primary determinant of SU content of purified human immunodeficiency virus type 1 and simian immunodeficiency virus. J. Virol. 76, 5315–5325 (2002).
Panico, M. et al. Mapping the complete glycoproteome of virion-derived HIV-1 gp120 provides insights into broadly neutralizing antibody binding. Sci. Rep. 6, 32956 (2016).
Moyo, T. et al. Molecular basis of unusually high neutralization resistance in tier 3 HIV-1 strain 253-11. J. Virol. 92, e02261-17 (2018).
Schoofs, T. et al. Broad and potent neutralizing antibodies recognize the silent face of the HIV envelope. Immunity 50, 1513–1529 (2019).
Escolano, A. et al. Immunization expands B cells specific to HIV-1 V3 glycan in mice and macaques. Nature 570, 468–473 (2019).
Schommers, P. et al. Restriction of HIV-1 escape by a highly broad and potent neutralizing antibody. Cell 180, 471–489 (2020).
Henderson, R. et al. Disruption of the HIV-1 envelope allosteric network blocks CD4-induced rearrangements. Nat. Commun. 11, 520 (2020).
Gristick, H. B. et al. Natively glycosylated HIV-1 Env structure reveals new mode for antibody recognition of the CD4-binding site. Nat. Struct. Mol. Biol. 23, 906–915 (2016).
Stadtmueller, B. M. et al. DEER spectroscopy measurements reveal multiple conformations of HIV-1 SOSIP envelopes that show similarities with envelopes on native virions. Immunity 49, 235–246 (2018).
Sattentau, Q. J. HIV gp120: double lock strategy foils host defences. Structure 6, 945–949 (1998).
Del Prete, G. Q. et al. Derivation and characterization of a simian immunodeficiency virus SIVmac239 variant with tropism for CXCR4. J. Virol. 83, 9911–9922 (2009).
Bess, J. W.Jr., Gorelick, R. J., Bosche, W. J., Henderson, L. E. & Arthur, L. O. Microvesicles are a source of contaminating cellular proteins found in purified HIV-1 preparations. Virology 230, 134–144 (1997).
Arthur, L. O. et al. Chemical inactivation of retroviral infectivity by targeting nucleocapsid protein zinc fingers: a candidate SIV vaccine. AIDS Res. Hum. Retroviruses 14(Suppl. 3), S311–319 (1998).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Kremer, J. R., Mastronarde, D. N. & McIntosh, J. R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71–76 (1996).
Mastronarde, D. N. & Held, S. R. Automated tilt series alignment and tomographic reconstruction in IMOD. J. Struct. Biol. 197, 102–113 (2017).
Agulleiro, J. I. & Fernandez, J. J. Tomo3D 2.0-Exploitation of Advanced Vector eXtensions (AVX) for 3D reconstruction. J. Struct. Biol. 189, 147–152 (2015).
Winkler, H. 3D reconstruction and processing of volumetric data in cryo-electron tomography. J. Struct. Biol. 157, 126–137 (2007).
Winkler, H. et al. Tomographic subvolume alignment and subvolume classification applied to myosin V and SIV envelope spikes. J. Struct. Biol. 165, 64–77 (2009).
Liu, J., Wright, E. R. & Winkler, H. 3D visualization of HIV virions by cryoelectron tomography. Methods Enzymol. 483, 267–290 (2010).
Kucukelbir, A., Sigworth, F. J. & Tagare, H. D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014).
Bordoli, L. & Schwede, T. Automated protein structure modeling with SWISS-MODEL workspace and the protein model portal. Methods Mol. Biol. 857, 107–136 (2012).
Pettersen, E. F. et al. UCSF chimera — a visualization system for exploratory research and analysis. J. Comp. Chem. 25, 1605–1612 (2004).
Afonine, P. V. et al. New tools for the analysis and validation of cryo-EM maps and atomic models. Acta Crystallogr. D Struct. Biol. 74, 814–840 (2018).
Goddard, T. D. et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018).
Juette, M. F. et al. Single-molecule imaging of non-equilibrium molecular ensembles on the millisecond timescale. Nat. Methods 13, 341–344 (2016).
Acknowledgements
This work was supported by the NIH grants RO1AI150560 to W.M., J.L. and S.C.B.; RO1 GM098859 to S.C.B.; PO1 AI150471 to W.M.; by a ViiV research grant to W.M. and J.L.; by an AmfAR (The Foundation for AIDS Research) grant, 109998-67-RKVA, to M.L.; by a grant from National Natural Science Foundation of China (No.31630002) to Z.L.; by the International AIDS Vaccine Initiative’s (IAVI’s) Neutralizing Antibody Consortium to P.D.K.; by the Intramural Research Program of the Vaccine Research Center (NIAID, NIH) to P.D.K.; and by contracts 75N91019D00054 and HHSN261200800001E from the National Cancer Institute, National Institutes of Health, to J.B. and J.D.L. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the US government.
Author information
Authors and Affiliations
Contributions
W.M., J.L. and P.D.K. conceived the project. W.L. performed cryo-ET experiments and data analysis. Z.L. performed data analysis and modeling, and wrote the manuscript draft. M.L. performed smFRET experiments and analysis. J.B. and J.D.L. provided BAL virions with high Env trimer content. C.W.C. and B.Z. provided sCD4D1D2, 17b, 1010-74 and 3BNC117 (Fabs). J.G. and T.Z. contributed structural analysis. D.S.T. and S.C.B. contributed smFRET technologies and software. J.L., P.D.K. and W.M. supervised all work. Z.L, P.D.K., W.M. and J.L. prepared the manuscript with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
S.C.B. holds an equity interest in Lumidyne Technologies. S.C.B. and W.M. hold the patent US 959385324 B2. W.M. and J.L. are recipients of a ViiV research grant.
Additional information
Peer review information Inês Chen was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Comparison of cryo-ET maps of the ligand-free HIV-1/SIV Env trimer.
a, In situ structure of Env trimer on the surface of SIV mac329 virus with truncated cytoplasmic tail (EMD 1246). b, Structure of Env trimer of SIVmneE11S virus (EMD 1216). c, Structure of Env trimer of HIV-1BaL virus (EMD 5019). d, Structure of ligand-free HIV-1BaL Env trimer (current work).
Extended Data Fig. 2 Cryo-ET resolution estimation.
a, Resolution estimation using FSC 0.5 as cutoff value. b, Local resolution estimation of ligand-free, 10-1074 and 3BNC117 double antibodies, and sCD4-17b bound HIV-1BaL Env trimer is estimated with Resmap. For each map, two projections are shown: Z plane projection is shown as a top view, Y plane projection is shown as a side view.
Extended Data Fig. 4 Visualization of variable loops and glycosylation sites on the top of the ligand-free Env trimer map.
a, Variable loop regions on the top of the Env trimer are differently colored in the map. b, Potential glycosylation densities at the apical surface of BaL strain virus. Ligand-free monomer of fully glycosylated JR-FL∆CT (PDB 5FUU) was docked in the cryo-ET map. The glycan moiety was shown as sticks.
Extended Data Fig. 5 Comparison between the cryo-ET model for HIV-1BaL Env and BG505 SOSIP.664 in complex with different antibody combinations.
Each ligand and its binding protomer were projected into central plane and the dihedral angles were measured with UCSF Chimera. a, Cryo-ET model of 10-1074-3 and BNC117 bound Env. b, 10-1074 bound BG505 SOSIP-based immunogen RC1 (PDB 6ORN). c, 3BNC117 bound BG505 SOSIP.664 (PDB 5V8M). d, 10-1074 and IOMA bound BG505 SOSIP.664 (PDB 5T3Z). All angular values are listed in panel (e).
Extended Data Fig. 6 10-1074 and 3BNC117 cannot stabilize State 1 of AT-2 treated HIV-1BaL Env.
a, b, 10-1074 and 3BNC117 Fabs exhibit a preference for State 1 of virus Env in the absence of AT2-inactivation (a) Representative fluorescence (top, donor Cy3 in green and acceptor Cy5 in red) and FRET trace (bottom, resulting FRET in blue and hidden Markov model idealization in red) of individual V1V4 labeled ligand-free HIV-1NL4-3 virus Env. (b) FRET histogram of HIV-1NL4-3 Env in the presence of broadly neutralization antibodies 10-1074 (50 μg ml−1) and 3BNC117 (50 μg ml−1), overlaid with that of ligand-free HIV-1NL4-3 Env. c, d, 10-1074 and 3BNC117 are unable to restore State 1-predominance observed on native virus Env after AT-2 treatment shifted the conformational equilibrium towards State 2. (c, d) experiments as in (a, b) of AT-2 chemically inactivated HIV-1NL4-3 virus Env, respectively. e, Quantification of relative state occupancy of HIV-1NL4-3 virus Env, derived from FRET histograms in (b, d).
Extended Data Fig. 7 The conformational effect of AT-2 lies in the ectodomain.
FRET histograms of HIV-1JR-FL Env without cytoplasmic tail under conditions: ligand-free and untreated (a); ligand-free Env after AT-2 treatment (b); and in the additional presence of 10-1074 (50 μg ml−1) and 3BNC117 (50 μg ml−1) (c). d, Quantification of the corresponding relative state occupancies.
Supplementary information
Rights and permissions
About this article
Cite this article
Li, Z., Li, W., Lu, M. et al. Subnanometer structures of HIV-1 envelope trimers on aldrithiol-2-inactivated virus particles. Nat Struct Mol Biol 27, 726–734 (2020). https://doi.org/10.1038/s41594-020-0452-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-020-0452-2