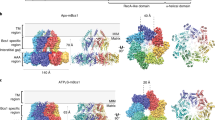
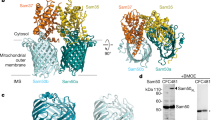
Abstract
Some proteins require completion of folding before translocation across a membrane into another cellular compartment. Yet the permeability barrier of the membrane should not be compromised and mechanisms have remained mostly elusive. Here, we present the structure of Saccharomyces cerevisiae Bcs1, an AAA-ATPase of the inner mitochondrial membrane. Bcs1 facilitates the translocation of the Rieske protein, Rip1, which requires folding and incorporation of a 2Fe–2S cluster before translocation and subsequent integration into the bc1 complex. Surprisingly, Bcs1 assembles into exclusively heptameric homo-oligomers, with each protomer consisting of an amphipathic transmembrane helix, a middle domain and an ATPase domain. Together they form two aqueous vestibules, the first being accessible from the mitochondrial matrix and the second positioned in the inner membrane, with both separated by the seal-forming middle domain. On the basis of this unique architecture, we propose an airlock-like translocation mechanism for folded Rip1.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The cryo-EM density maps and corresponding atomic models reported in this paper have been deposited in the Electron Microscopy Data Bank and Protein Data Bank with the accession codes EMD-10192, EMD-10193 and EMD-10194, as well as PDB 6SH3, 6SH4 and 6SH5, for the Bcs1-ADP, Bcs1-Apo1 and Bcs1-Apo2 states, respectively.
References
Rapoport, T. A., Li, L. & Park, E. Structural and mechanistic insights into protein translocation. Annu. Rev. Cell Dev. Biol. 33, 369–390 (2017).
Green, E. R. & Mecsas, J. Bacterial secretion systems: an overview. Microbiol. Spectr. 4, VMBF-0012-2015 (2016).
Berks, B. C. The twin-arginine protein translocation pathway. Annu. Rev. Biochem. 84, 843–864 (2015).
Hunte, C., Koepke, J., Lange, C., Rossmanith, T. & Michel, H. Structure at 2.3 Å resolution of the cytochrome bc(1) complex from the yeast Saccharomyces cerevisiae co-crystallized with an antibody Fv fragment. Structure 8, 669–684 (2000).
Kispal, G., Csere, P., Guiard, B. & Lill, R. The ABC transporter Atm1p is required for mitochondrial iron homeostasis. FEBS Lett. 418, 346–350 (1997).
Kispal, G., Csere, P., Prohl, C. & Lill, R. The mitochondrial proteins Atm1p and Nfs1p are essential for biogenesis of cytosolic Fe/S proteins. EMBO J. 18, 3981–3989 (1999).
Wagener, N., Ackermann, M., Funes, S. & Neupert, W. A pathway of protein translocation in mitochondria mediated by the AAA-ATPase Bcs1. Mol. Cell 44, 191–202 (2011).
Molik, S., Karnauchov, I., Weidlich, C., Herrmann, R. G. & Klosgen, R. B. The Rieske Fe/S protein of the cytochrome b6/f complex in chloroplasts: missing link in the evolution of protein transport pathways in chloroplasts? J. Biol. Chem. 276, 42761–42766 (2001).
Aldridge, C., Spence, E., Kirkilionis, M. A., Frigerio, L. & Robinson, C. Tat-dependent targeting of Rieske iron–sulphur proteins to both the plasma and thylakoid membranes in the cyanobacterium Synechocystis PCC6803. Mol. Microbiol. 70, 140–150 (2008).
Nobrega, F. G., Nobrega, M. P. & Tzagoloff, A. BCS1, a novel gene required for the expression of functional Rieske iron-sulfur protein in Saccharomyces cerevisiae. EMBO J. 11, 3821–3829 (1992).
Cruciat, C. M., Hell, K., Folsch, H., Neupert, W. & Stuart, R. A. Bcs1p, an AAA-family member, is a chaperone for the assembly of the cytochrome bc(1) complex. EMBO J. 18, 5226–5233 (1999).
Frickey, T. & Lupas, A. N. Phylogenetic analysis of AAA proteins. J. Struct. Biol. 146, 2–10 (2004).
Sauer, R. T. et al. Sculpting the proteome with AAA+ proteases and disassembly machines. Cell 119, 9–18 (2004).
Truscott, K. N., Lowth, B. R., Strack, P. R. & Dougan, D. A. Diverse functions of mitochondrial AAA+ proteins: protein activation, disaggregation, and degradation. Biochem. Cell Biol. 88, 97–108 (2010).
Zolkiewski, M. A camel passes through the eye of a needle: protein unfolding activity of Clp ATPases. Mol. Microbiol. 61, 1094–1100 (2006).
Tatsuta, T. & Langer, T. AAA proteases in mitochondria: diverse functions of membrane-bound proteolytic machines. Res. Microbiol. 160, 711–717 (2009).
Chen, Y. C. et al. Msp1/ATAD1 maintains mitochondrial function by facilitating the degradation of mislocalized tail-anchored proteins. EMBO J. 33, 1548–1564 (2014).
Okreglak, V. & Walter, P. The conserved AAA-ATPase Msp1 confers organelle specificity to tail-anchored proteins. Proc. Natl Acad. Sci. USA 111, 8019–8024 (2014).
Wohlever, M. L., Mateja, A., McGilvray, P. T., Day, K. J. & Keenan, R. J. Msp1 is a membrane protein dislocase for tail-anchored proteins. Mol. Cell 67, 194–202 (2017).
Puchades, C. et al. Structure of the mitochondrial inner membrane AAA+ protease YME1 gives insight into substrate processing. Science 358, eaao0464 (2017).
Lee, S. et al. Cryo-EM structures of the Hsp104 protein disaggregase captured in the ATP conformation. Cell Rep. 26, 29–36.e3 (2019).
Twomey, E. C. et al. Substrate processing by the Cdc48 ATPase complex is initiated by ubiquitin unfolding. Science 365, eaax1033 (2019).
Majumder, P. et al. Cryo-EM structures of the archaeal PAN-proteasome reveal an around-the-ring ATPase cycle. Proc. Natl Acad. Sci. USA 116, 534–539 (2019).
White, K. I., Zhao, M., Choi, U. B., Pfuetzner, R. A. & Brunger, A. T. Structural principles of SNARE complex recognition by the AAA+ protein NSF. Elife 7, e38888 (2018).
Folsch, H., Guiard, B., Neupert, W. & Stuart, R. A. Internal targeting signal of the BCS1 protein: a novel mechanism of import into mitochondria. EMBO J. 15, 479–487 (1996).
Wagener, N. & Neupert, W. Bcs1, a AAA protein of the mitochondria with a role in the biogenesis of the respiratory chain. J. Struct. Biol. 179, 121–125 (2012).
Miller, J. M. & Enemark, E. J. Fundamental characteristics of AAA+ protein family structure and function. Archaea 2016, 9294307 (2016).
Nouet, C., Truan, G., Mathieu, L. & Dujardin, G. Functional analysis of yeast bcs1 mutants highlights the role of Bcs1p-specific amino acids in the AAA domain. J. Mol. Biol. 388, 252–261 (2009).
Atkinson, A. et al. The LYR protein Mzm1 functions in the insertion of the Rieske Fe/S protein in yeast mitochondria. Mol. Cell. Biol. 31, 3988–3996 (2011).
de la Pena, A. H., Goodall, E. A., Gates, S. N., Lander, G. C. & Martin, A. Substrate-engaged 26S proteasome structures reveal mechanisms for ATP-hydrolysis-driven translocation. Science 362, eaav0725 (2018).
Monroe, N., Han, H., Shen, P. S., Sundquist, W. I. & Hill, C. P. Structural basis of protein translocation by the Vps4-Vta1 AAA ATPase. Elife 6, e24487 (2017).
Puchades, C. et al. Unique structural features of the mitochondrial AAA+ protease AFG3L2 reveal the molecular basis for activity in health and disease. Mol. Cell 75, 1073–1085.e6 (2019).
Kyte, J. & Doolittle, R. F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132 (1982).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Su, M. et al. Mechanism of Vps4 hexamer function revealed by cryo-EM. Sci. Adv. 3, e1700325 (2017).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7, e42166 (2018).
Fernandez-Leiro, R. & Scheres, S. H. W. A pipeline approach to single-particle processing in RELION. Acta Crystallogr. D Struct. Biol. 73, 496–502 (2017).
Hildebrand, A., Remmert, M., Biegert, A. & Soding, J. Fast and accurate automatic structure prediction with HHpred. Proteins 77, 128–132 (2009).
Webb, B. & Sali, A. Protein structure modeling with MODELLER. Methods Mol. Biol. 1654, 39–54 (2017).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Kidmose, R. T. et al. Namdinator – automatic molecular dynamics flexible fitting of structural models into cryo-EM and crystallography experimental maps. IUCrJ 6, 526–531 (2019).
Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Struct. Biol. 74, 531–544 (2018).
Goddard, T. D. et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018).
Acknowledgements
We thank S. Rieder, C. Ungewickell and S. Esser for technical assistance and help with experiments and M. Ameismeier for support with electron microscopy. This work was supported by funding from the Deutsche Forschungsgemeinschaft (DFG) through the GRK1721 to R.B., a DFG fellowship through the QBM (Quantitative Biosciences Munich) graduate school to L.K. and the DFG grant WA3802/1-1 and the LMUexcellent program of the Bundesexzellenzinitiative to N.W. and W.N.
Author information
Authors and Affiliations
Contributions
R.B. and W.N. designed the study and L.K., N.W., T.B. and R.B. wrote the manuscript. N.W. purified and characterized Bcs1 complexes, L.K. prepared cryo-EM samples and, together with O.B., collected data. L.K. processed cryo-EM maps, built molecular models and, together with T.B. and R.B., interpreted the structures.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Anke Sparmann was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 SDS-PAGE gel of purified Bcs1.
The concentrated Bcs1-His eluate (see Materials and Methods) was loaded on a 4-10% gradient gel and bands were analyzed by mass spectrometry. This mild and fast one-step affinity purification yields Bcs1 assemblies active in nucleotide and substrate binding7; remaining background proteins represent abundant mitochondrial proteins that unspecifically interact with Ni-NTA resin.
Extended Data Fig. 2 Cryo-EM data processing of the Bcs1 complex with and without ADP.
(a) Representative original micrograph and 2D class averages for the ADP (left) and the Apo (right) datasets. (b) “Gold standard” Fourier-Shell correlation (FSC) curves when applying C1 and C7 symmetry during refinement. The final resolutions were 3.4 Å (C7) and 4.0 Å (C1). (c) Final C1 and C7 symmetrized maps filtered and colored according to local resolution as calculated with RELION-3.
Extended Data Fig. 3 3D classification scheme for the Bcs1 apo data set.
The classification regiment is described in detail in the Methods section. In brief, after import and CTF-estimation, particles were first picked using the structure of the Vps4 AAA-ATPase hexamer as a template (EMD-8588)37. Particles were 2D classified and an ab initio model was obtained using cryoSPARC v.2. From the refined map, a better reference for RELION autopicking was obtained and resulting particles were again 2D classified. Good classes were subjected to 3D classification yielding one stable feature-containing class that could be sub-classified further. Two stable classes were then 3D refined using either C1 or C7 symmetry yielding the final Bcs1 Apo1 and Bcs1 Apo2 maps. K: number of classes.
Extended Data Fig. 4 3D classification scheme for the Bcs1 ADP dataset.
The classification regiment is described in detail in the Methods section. In brief, autopicking in RELION-3 (Laplacian of Gaussian (LoG) method)38 was performed using the previously obtained structure of Bcs1 Apo1 as a reference. 2D classification and 3D refinement were performed and auto-picking repeated using the refined map to optimize the quality and yield of good Bcs1 complex particles. After a second round of 2D classification and refinement, 3D classification yielded in one class that could be refined including per particle CTF correction and beam tilt correction in RELION-3 using C1 and C7 symmetry to a final resolution of and 3.4 Å and 4.0 Å, respectively. K: number of classes.
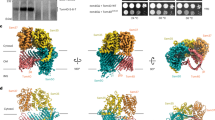
Extended Data Fig. 5 Comparison of two membrane bound mitochondrial AAA proteins Yme1 and Bcs1.
Shown are two protomers of both Yme1 (a) (PDB: 6AZ0)20 and Bcs1 either in the ADP state (b) or the Apo1 state (c). The pore loops and preceding beta strands are shown in orange (PL1, β2) and blue (PL2, β3). The six AAA domains of Yme1 form a spiral staircase configuration. Displayed here are the ADP-bound (purple) and Apo (cyan) protomers, which constitute the bottom and top end of this spiral. The two pore loops of every AAA domain protrude towards the central axis of this complex, where they can interact with the peptide backbone of the substrate (red) via the conserved tyrosine residues. In this configuration, the beta strands β2 and β3, which precede the two pore loops point towards the central axis. Conversely, in Bcs1, these loops and the corresponding preceding beta strands do not point towards the opening of the ring of AAA domains, but rather point away from the ring plane in both displayed conformations. In the ADP state these loops are oriented approximately parallel to the central axis, in the apo states they are slightly tilted towards the central axis.
Supplementary information
Supplementary Video 1
Overview of Bcs1 and transitions between the Bcs1 ADP and apo states.
Rights and permissions
About this article
Cite this article
Kater, L., Wagener, N., Berninghausen, O. et al. Structure of the Bcs1 AAA-ATPase suggests an airlock-like translocation mechanism for folded proteins. Nat Struct Mol Biol 27, 142–149 (2020). https://doi.org/10.1038/s41594-019-0364-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-019-0364-1
This article is cited by
-
A concerted ATPase cycle of the protein transporter AAA-ATPase Bcs1
Nature Communications (2023)
-
Visualizing maturation factor extraction from the nascent ribosome by the AAA-ATPase Drg1
Nature Structural & Molecular Biology (2022)
-
Cryo-EM structure of transmembrane AAA+ protease FtsH in the ADP state
Communications Biology (2022)
-
Modelling of BCS1L-related human mitochondrial disease in Drosophila melanogaster
Journal of Molecular Medicine (2021)
-
Rearranging AAA+ architecture to accommodate folded substrates
Nature Structural & Molecular Biology (2020)