Abstract
The decoration of secretory glycoproteins and glycolipids with sialic acid is critical to many physiological and pathological processes. Sialyation is dependent on a continuous supply of sialic acid into Golgi organelles in the form of CMP-sialic acid. Translocation of CMP-sialic acid into Golgi is carried out by the CMP-sialic acid transporter (CST). Mutations in human CST are linked to glycosylation disorders, and CST is important for glycopathway engineering, as it is critical for sialyation efficiency of therapeutic glycoproteins. The mechanism of how CMP-sialic acid is recognized and translocated across Golgi membranes in exchange for CMP is poorly understood. Here we have determined the crystal structure of a Zea mays CST in complex with CMP. We conclude that the specificity of CST for CMP-sialic acid is established by the recognition of the nucleotide CMP to such an extent that they are mechanistically capable of both passive and coupled antiporter activity.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The coordinates and the structure factors for CSTZM have been deposited in the Protein Data Bank with accession numbers PDB 6I1R (CMP-bound CSTZM structure) and PDB 6I1Z (apo CSTZM structure). Data supporting the findings of this manuscript and requests for resources and reagents are available upon reasonable request to D.D. (ddrew@dbb.su.se).
References
Varki, A. Sialic acids in human health and disease. Trends Mol. Med. 14, 351–360 (2008).
Varki, A. & Schauer, R. in Essentials of Glycobiology 2nd edn (eds Varki, A., Cummings, R. D. & Esko, J. D. et al.) Ch. 14 (Cold Spring Harbor Laboratory Press, 2009).
Yoo, S. W. et al. Sialylation regulates brain structure and function. FASEB J. 29, 3040–3053 (2015).
Han, J. et al. Genome-wide CRISPR/Cas9 screen identifies host factors essential for influenza virus replication. Cell Rep. 23, 596–607 (2018).
Bull, C., Stoel, M. A., den Brok, M. H. & Adema, G. J. Sialic acids sweeten a tumor’s life. Cancer Res. 74, 3199–3204 (2014).
Rodrigues, E. & Macauley, M. S. Hypersialylation in cancer: modulation of inflammation and therapeutic opportunities.Cancers (Basel). 10(), 207 (2018).
Bauer, J. & Osborn, H. M. Sialic acids in biological and therapeutic processes: opportunities and challenges. Future Med. Chem. 7, 2285–2299 (2015).
Hadley, B. et al. Structure and function of nucleotide sugar transporters: current progress. Comput. Struct. Biotechnol. J. 10, 23–32 (2014).
Eckhardt, M., Muhlenhoff, M., Bethe, A. & Gerardy-Schahn, R. Expression cloning of the Golgi CMP-sialic acid transporter. Proc. Natl Acad. Sci. USA 93, 7572–7576 (1996).
Hirschberg, C. B., Robbins, P. W. & Abeijon, C. Transporters of nucleotide sugars, ATP, and nucleotide sulfate in the endoplasmic reticulum and Golgi apparatus. Annu. Rev. Biochem 67, 49–69 (1998).
Wex, B. et al. SLC35B4, an inhibitor of gluconeogenesis, responds to glucose stimulation and downregulates Hsp60 among other proteins in HepG2 liver cell lines. Molecules 23, E1350 (2018).
Yazbek, S. N. et al. Deep congenic analysis identifies many strong, context-dependent QTLs, one of which, Slc35b4, regulates obesity and glucose homeostasis. Genome Res. 21, 1065–1073 (2011).
Winter, G. E. et al. The solute carrier SLC35F2 enables YM155-mediated DNA damage toxicity. Nat. Chem. Biol. 10, 768–773 (2014).
Rowland, A., Mackenzie, P. I. & Miners, J. O. Transporter-mediated uptake of UDP-glucuronic acid by human liver microsomes: assay conditions, kinetics, and inhibition. Drug Metab. Dispos. 43, 147–153 (2015).
Kutsuno, Y., Itoh, T., Tukey, R. H. & Fujiwara, R. Glucuronidation of drugs and drug-induced toxicity in humanized UDP-glucuronosyltransferase 1 mice. Drug Metab. Dispos. 42, 1146–1152 (2014).
Caffaro, C. E. et al. A single Caenorhabditis elegans Golgi apparatus-type transporter of UDP-glucose, UDP-galactose, UDP-N-acetylglucosamine, and UDP-N-acetylgalactosamine. Biochemistry 47, 4337–4344 (2008).
Berninsone, P., Eckhardt, M., Gerardy-Schahn, R. & Hirschberg, C. B. Functional expression of the murine Golgi CMP-sialic acid transporter in Saccharomyces cerevisiae. J. Biol. Chem. 272, 12616–12619 (1997).
Ng, B. G. et al. Encephalopathy caused by novel mutations in the CMP-sialic acid transporter, SLC35A1. Am. J. Med. Genet. A 173, 2906–2911 (2017).
Martinez-Duncker, I. et al. Genetic complementation reveals a novel human congenital disorder of glycosylation of type II, due to inactivation of the Golgi CMP-sialic acid transporter. Blood 105, 2671–2676 (2005).
Pagan, J. D., Kitaoka, M. & Anthony, R. M. Engineered sialylation of pathogenic antibodies in vivo attenuates autoimmune disease. Cell 172, 564–577 e13 (2018).
Ghaderi, D., Taylor, R. E., Padler-Karavani, V., Diaz, S. & Varki, A. Implications of the presence of N-glycolylneuraminic acid in recombinant therapeutic glycoproteins. Nat. Biotechnol. 28, 863–867 (2010).
Kwak, C. Y. et al. Enhancing the sialylation of recombinant EPO produced in CHO cells via the inhibition of glycosphingolipid biosynthesis. Sci. Rep. 7, 13059 (2017).
Parker, J. L. & Newstead, S. Structural basis of nucleotide sugar transport across the Golgi membrane. Nature 551, 521–524 (2017).
Perez, M. & Hirschberg, C. B. Topography of glycosylation reactions in the rough endoplasmic reticulum membrane. J. Biol. Chem. 261, 6822–6830 (1986).
Martinez-Duncker, I., Mollicone, R., Codogno, P. & Oriol, R. The nucleotide-sugar transporter family: a phylogenetic approach. Biochimie 85, 245–260 (2003).
Bakker, H. et al. A CMP-sialic acid transporter cloned from Arabidopsis thaliana. Carbohydr. Res. 343, 2148–2152 (2008).
Takashima, S. et al. Analysis of CMP-sialic acid transporter-like proteins in plants. Phytochemistry 70, 1973–1981 (2009).
Drew, D. et al. GFP-based optimization scheme for the overexpression and purification of eukaryotic membrane proteins in Saccharomyces cerevisiae. Nat. Protoc. 3, 784–798 (2008).
Kawate, T. & Gouaux, E. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure 14, 673–681 (2006).
Nji, E., Chatzikyriakidou, Y., Landreh, M. & Drew, D. An engineered thermal-shift screen reveals specific lipid preferences of eukaryotic and prokaryotic membrane proteins. Nat. Commun. 9, 4253 (2018).
Milla, M. E. & Hirschberg, C. B. Reconstitution of Golgi vesicle CMP-sialic acid and adenosine 3’-phosphate 5’-phosphosulfate transport into proteoliposomes. Proc. Natl Acad. Sci. USA 86, 1786–1790 (1989).
Tsuchiya, H. et al. Structural basis for amino acid export by DMT superfamily transporter YddG. Nature 534, 417–420 (2016).
Lee, Y. et al. Structure of the triose-phosphate/phosphate translocator reveals the basis of substrate specificity. Nat. Plants 3, 825–832 (2017).
Takemoto, M., Lee, Y., Ishitani, R. & Nureki, O. Free energy landscape for the entire transport cycle of triose-phosphate/phosphate translocator. Structure 26, 1284–1296.e4 (2018).
Mohamed, M. et al. Intellectual disability and bleeding diathesis due to deficient CMP–sialic acid transport. Neurology 81, 681–687 (2013).
Song, Z. Roles of the nucleotide sugar transporters (SLC35 family) in health and disease. Mol. Asp. Med. 34, 590–600 (2013).
Carey, D. J., Sommers, L. W. & Hirschberg, C. B. CMP-N-acetylneuraminic acid: isolation from and penetration into mouse liver microsomes. Cell 19, 597–605 (1980).
Wahlgren, W. Y. et al. Substrate-bound outward-open structure of a Na+-coupled sialic acid symporter reveals a new Na+ site. Nat. Commun. 9, 1753 (2018).
Aoki, K., Ishida, N. & Kawakita, M. Substrate recognition by nucleotide sugar transporters: further characterization of substrate recognition regions by analyses of UDP-galactose/CMP-sialic acid transporter chimeras and biochemical analysis of the substrate specificity of parental and chimeric transporters. J. Biol. Chem. 278, 22887–22893 (2003).
Varki, A. Colloquium paper: uniquely human evolution of sialic acid genetics and biology. Proc. Natl Acad. Sci. USA 107 (Suppl 2), 8939–8946 (2010).
Seveno, M. et al. Glycoprotein sialylation in plants? Nat. Biotechnol. 22, 1351–1352 (2004); author reply 22, 1352–1353.
Shah, M. M., Fujiyama, K., Flynn, C. R. & Joshi, L. Sialylated endogenous glycoconjugates in plant cells. Nat. Biotechnol. 21, 1470–1471 (2003).
Nakagawa, T., Shimada, Y., Pavlova, N. V., Li, S. C. & Li, Y. T. Cloning and expression of 3-deoxy-d-manno-oct-2-ulosonic acid alpha-ketoside hydrolase from oyster hepatopancreasdagger. Glycobiology 25, 1431–1440 (2015).
Caffaro, C. E., Hirschberg, C. B. & Berninsone, P. M. Independent and simultaneous translocation of two substrates by a nucleotide sugar transporter. Proc. Natl Acad. Sci. USA 103, 16176–16181 (2006).
Berninsone, P., Hwang, H. Y., Zemtseva, I., Horvitz, H. R. & Hirschberg, C. B. SQV-7, a protein involved in Caenorhabditis elegans epithelial invagination and early embryogenesis, transports UDP-glucuronic acid, UDP-N-acetylgalactosamine, and UDP-galactose. Proc. Natl Acad. Sci. USA 98, 3738–3743 (2001).
Drew, D. & Boudker, O. Shared molecular mechanisms of membrane transporters. Annu. Rev. Biochem. 85, 543–572 (2016).
Aoki, K., Ishida, N. & Kawakita, M. Substrate recognition by UDP-galactose and CMP-sialic acid transporters. Different sets of transmembrane helices are utilized for the specific recognition of UDP-galactose and CMP-sialic acid. J. Biol. Chem. 276, 21555–21561 (2001).
Weis, W. I. & Kobilka, B. K. The molecular basis of G protein–coupled receptor activation. Annu. Rev. Biochem. 87, 897–919 (2018).
Chiaramonte, M. et al. Inhibition of CMP-sialic acid transport into Golgi vesicles by nucleoside monophosphates. Biochemistry 40, 14260–14267 (2001).
Caffaro, C. E. & Hirschberg, C. B. Nucleotide sugar transporters of the Golgi apparatus: from basic science to diseases. Acc. Chem. Res 39, 805–812 (2006).
Schapiro, F. B. & Grinstein, S. Determinants of the pH of the Golgi complex. J. Biol. Chem. 275, 21025–21032 (2000).
Al-Saraireh, Y. M. et al. Pharmacological inhibition of polysialyltransferase ST8SiaII modulates tumour cell migration. PLoS One 8, e73366 (2013).
Miyazaki, T., Angata, K., Seeberger, P. H., Hindsgaul, O. & Fukuda, M. CMP substitutions preferentially inhibit polysialic acid synthesis. Glycobiology 18, 187–194 (2008).
Kean, E. L., Munster-Kuhnel, A. K. & Gerardy-Schahn, R. CMP-sialic acid synthetase of the nucleus. Biochim. Biophys. Acta 1673, 56–65 (2004).
Eckhardt, M., Gotza, B. & Gerardy-Schahn, R. Mutants of the CMP-sialic acid transporter causing the Lec2 phenotype. J. Biol. Chem. 273, 20189–20195 (1998).
Takeshima-Futagami, T. et al. Amino acid residues important for CMP-sialic acid recognition by the CMP-sialic acid transporter: analysis of the substrate specificity of UDP-galactose/CMP-sialic acid transporter chimeras. Glycobiology 22, 1731–1740 (2012).
Chan, K. F., Zhang, P. & Song, Z. Identification of essential amino acid residues in the hydrophilic loop regions of the CMP-sialic acid transporter and UDP-galactose transporter. Glycobiology 20, 689–701 (2010).
Forrest, L. R. Structural symmetry in membrane proteins. Annu. Rev. Biophys. 44, 311–337 (2015).
Vergara-Jaque, A., Fenollar-Ferrer, C., Kaufmann, D. & Forrest, L. R. Repeat-swap homology modeling of secondary active transporters: updated protocol and prediction of elevator-type mechanisms. Front. Pharm. 6, 183 (2015).
Henderson, R. K., Fendler, K. & Poolman, B. Coupling efficiency of secondary active transporters. Curr. Opin. Biotechnol. 58, 62–71 (2018).
Kota, J., Gilstring, C. F. & Ljungdahl, P. O. Membrane chaperone Shr3 assists in folding amino acid permeases preventing precocious ERAD. J. Cell Biol. 176, 617–628 (2007).
Newstead, S., Kim, H., von Heijne, G., Iwata, S. & Drew, D. High-throughput fluorescent-based optimization of eukaryotic membrane protein overexpression and purification in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA 104, 13936–13941 (2007).
Sonoda, Y. et al. Benchmarking membrane protein detergent stability for improving throughput of high-resolution X-ray structures. Structure 19, 17–25 (2011).
Hattori, M., Hibbs, R. E. & Gouaux, E. A fluorescence-detection size-exclusion chromatography-based thermostability assay for membrane protein precrystallization screening. Structure 20, 1293–1299 (2012).
Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Schroder, G. F., Levitt, M. & Brunger, A. T. Super-resolution biomolecular crystallography with low-resolution data. Nature 464, 1218–1222 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, (213–221 (2010).
Vagin, A. A. et al. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr. D Biol. Crystallogr. 60, 2184–2195 (2004).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Acknowledgements
We are grateful to Y. Chatzikyriakidou for generation of Fig. 1a and M. Claesson for assistance with data collection and manuscript preparation. We wish to especially thank A. McCarthy for extra assistance at the European Synchrotron Radiation Facility and the Diamond Light Source synchrotron beamline scientists for their excellent assistance. This work was funded by the Knut and Alice Wallenberg Foundation (to D.D.). D.D. acknowledges support from the Wenner-Gren foundation and EMBO through the Young Investigator Program (YIP).
Author information
Authors and Affiliations
Contributions
D.D. designed the project. Purification and crystallization of CSTZM was carried out by E.N. Data collection was carried out by E.N., M.C. and D.D. Structure determination and refinement of CSTZM was carried out by A.G. Binding experiments were carried out by E.N. and A.G. Transport assays were carried out by A.A.Q. The manuscript was prepared by D.D. and A.G. with assistance from E.N., A.A.Q. and M.C. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Fig. 1 Stability and substrate binding to CSTZM and hCST.
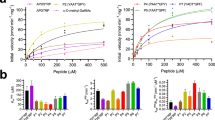
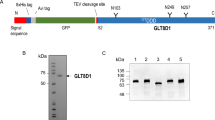
a, Size-exclusion chromatogram of CSTZM and corresponding SDS-PAGE-gel with DDM purified CSTZM migrating predominantly as a dimer. b, Determination of the Michaelis constant (KM) for [3H]-CMP/CMP-Sia exchange for CSTZM and hCST. Kinetic curves were fitted from data points recorded over a range of increasing CMP concentrations (internal liposomes containing steady-state levels of CMP-Sia) at 90 s time points and fitted by non-linear regression using data from 3 independent experiments (the values reported are the mean ± s.e.m. of the fit) c, The GFP-TS melting curves using GFP-fusions for CSTZM in crude-detergent solubilized membranes (black open circles) and for hCST (cyan filled squares); errors bars, s.e.m.; n = 3 and the values reported for the apparent Tm are the mean ± s.e.m. of the data fitting. d, The apparent Tm for CSTZM and hCST were determined as in c, and further in the presence of 1 mM CMP, 1 mM UMP, 1 mM GMP, 1 mM TMP, 10 mM CMP-sialic acid or 10 mM sialic acid. The difference in melting temperature ΔTm is shown for CSTZM (filled bars) and hCST (open bars); errors bars are the s.e.m. 3 independent experiments (cyan-filled circles). Markedly, absolute ΔTm differences between CMP and UMP are equivalent to that measured using the CPM assay to estimate binding differences between ATP and AMP in the ADP/ATP exchanger (Majd, H. et al., Elife. 7, 2018). e, FSEC traces of DDM-solubilised membranes of CSTZM and hCST constructs assessed for substrate-binding by GFP-TS (see Supplementary Table 1).
Supplementary Fig. 2 Superimposition of apo- and substrate-bound outward-facing CSTZM structures to Vrg4 and other structural homologues.
a, Representative portions of the 2mFo-DFc electron density map (1.4σ), which is shown here for the TM segments involved in CMP binding. b, Cartoon representation of outward-facing 2.8 Å resolution structure of CSTZM in complex with CMP (colored as in Fig. 1d) and the 3.6 Å outward-facing structure of Vrg4 in complex with GDP-mannose (PDB id: 5OGK; light grey), as viewed from the top of the membrane. The CMP and GDP-mannose moieties from CSTZM and Vrg4 respectively, are shown as sticks. The polder (OMIT) map (Liebschner, D. et al., Acta Crystallogr D Struct Biol. 73, 148-157, 2017) for CMP contoured at 3σ is also shown. c, As in b., the apo 3.4Å outward-facing structure of CSTZM (colored in wheat) and the apo 3.2 Å resolution outward-facing Vrg4 (PDB id: 50GE; light grey) structures. d, Structural comparison of CMP bound (in wheat) and apo (in light grey) CST. The structures superposed with a very low r.m.s.d. of 0.3 Å, indicating no significant conformation difference between the two isoforms. e, Cartoon representation of outward facing bacterial amino acid transporter YddG (PDB id: 5I20; light grey) and CSTZM (wheat). A lipid molecule (in grey sticks) is present at the putative binding pocket of YddG. Both the structures differ with an r.m.s.d. of 4.1 Å, owing to the large evolutionary gap between the two DMT superfamily members. f, Cartoon representation of outward open CSTZM and occluded model of plant triose-phosphate/phosphate translocator (PDB id: 5Y78; light grey). Large inward motion of helix 3 and 4 during transport and conformational switching is consistent with the observations made using structural inverted repeats of CSTZM (see Fig. 3a). Dimerization helices 5 and 10 are not shown for clarity.
Supplementary Fig. 3 Comparison of substrate-binding residues in CSTZM and Vrg4 are very different.
a, Cartoon representation illustrating the pronounced differences of the substrate-binding site residues when comparing the outward-facing CSTZM structure in complex with CMP (colored cyan) and the outward-facing Vrg4 structure in complex with GDP-mannose (colored light-grey). The position of the bound CMP is shown as an oval and residues as sticks. b, Cartoon representation illustrating the pronounced differences of the substrate-binding site residues in the Vrg4 structure in complex with GDP-mannose (colored grey) when compared to the structure of CSTZM in complex with CMP (colored cyan). The position of the GDP-mannose is shown as an oval and residues as sticks. c, Slab through the outward-facing CSTZM as viewed within the plane of membrane with CMP (shown as green sticks). All cavity waters in CSTZM are shown as red spheres, which was generated by overlaying separately to the sliced surface. To visualise structural sequence conservation, the ConSurf server (Landau, M. et al., Nucleic Acids Res. 33, W299-302, 2005) was used with default settings (colored based on sequence conservation; low-cyan to high-dark-red). The sequence alignment for conservation analysis was generated using Clustal Omega by comparing 63 CSTZM homologues with sequence identity ranging from 23 to 91 %. d, Modelled positions and 2mFO-DFC electron density map (1.0 σ) for the observed waters and CMP in the CSTZM complex and the residues involved in water coordination. Residues and CMP are shown as sticks and waters as red spheres.
Supplementary Fig. 4 Binding and transport activities for CSTZM and hCST.
a, CMP binding to CSTZM using GFP-fusions as estimated by a shift in ΔTm after addition of 1 mM CMP to WT (open bars), and to mutants of residues not interacting with crystallographic waters (N106A and S267A) in the cavity, but are present in close proximity to the water network (filled bars). Residues W209 and N206 are located in the CMP binding pocket of CSTZM, but are not conserved in hCST. Non-specific binding was estimated with addition of UMP (red bars). b, The GFP-TS assay was used to determine binding of CMP to purified GFP-fusions of CSTZM (black squares) and hCST (cyan squares). Binding affinities (Kd) were calculated from data points recorded over a range of CMP concentrations, and these were fitted by non-linear regression using data from 3 independent experiments. As has been observed in other transporters (Shukla, S. et al., J Biol Chem. 292, 7066-7076, 2017), the absolute apparent binding affinities in detergent, although consistently different, were significantly weaker than IC50 and KM values determined in membranes. c, as in b for the CSTZM Ser79Ala and hCST Ser95Ala mutants shown to have enhanced CMP binding in (Fig. 2b, c). The binding affinities were estimated using the solubilized membranes. d, as in b for CMP-Sia; see Methods). e, Time-dependent uptake of [3H]-CMP by CSTZM (open circles). Non-specific uptake was estimated with empty liposomes (filled squares). f, Time-dependent uptake of [3H]-CMP by hCST (open circles). Non-specific uptake was estimated with empty liposomes (filled squares). In all experiments errors bars, s.e.m.; n = 3.
Supplementary Fig. 5 Modelled position of CMP-sialic acid and comparison to Vrg4 and TPT.
a, The substrate-binding site in the outward facing CSTZM structure with modelled CMP-sialic acid (TMs in light brown). The CMP-sialic acid is shown as green sticks, and residues coordinating the modelled CMP-sialic acid as sticks colored in rainbow (as in Fig. 1d). b, View of the CSTZM surface (colored according to the calculated electrostatic potential, presented in blue-positive to red-negative) as viewed from the top with the modelled CMP-sialic acid (shown as green sticks). c-e, Cartoon representation of the structural comparison of CMP bound CSTZM (brown), GDP-mannose bound Vrg4 (cyan; PDB id: 5OGK) and 3-Phosphoglyceric acid (3PG) TPT (pink; PDB id: 5Y78) showing the position of the substrate relative to structural conserved TM4- and TM9-embedded lysine residues (dashed lines represent direct electrostatic interactions between lysine and terminal phosphate moiety), f, Structures of the α-keto acids, mentioned in the text. 9-carbon α-keto acids; N-acetylneuraminic acid (Neu5Ac) and 2-keto-3-deoxy-d-glycero-d-galacto-nonulosonic acid (Kdn), with the 8-carbon α-keto acid 2-keto-3-deoxy-d-manno-octulosonic acid (Kdo).
Supplementary Fig. 6 Intracellular intra-bundle polar interactions.
Cartoon representation showing the residues which by polar interactions stabilize the two transport bundles (colored cyan and wheat, respectively) in the outward-facing conformation. CSTZM E181 is equivalent to hCST E196, which when mutated to lysine (E196K) causes a congenital disorder of glycosylation as it abolishes CST transport activity (Fig. 3d).
Supplementary information
Supplementary Information
Supplementary Figures 1–6, Supplementary Table 1, Supplementary Note
Rights and permissions
About this article
Cite this article
Nji, E., Gulati, A., Qureshi, A.A. et al. Structural basis for the delivery of activated sialic acid into Golgi for sialyation. Nat Struct Mol Biol 26, 415–423 (2019). https://doi.org/10.1038/s41594-019-0225-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-019-0225-y
This article is cited by
-
The GFP thermal shift assay for screening ligand and lipid interactions to solute carrier transporters
Nature Protocols (2021)
-
The structural basis of promiscuity in small multidrug resistance transporters
Nature Communications (2020)
-
Structure and drug resistance of the Plasmodium falciparum transporter PfCRT
Nature (2019)
-
Structural basis for substrate specificity and regulation of nucleotide sugar transporters in the lipid bilayer
Nature Communications (2019)