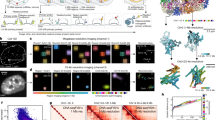
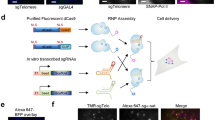
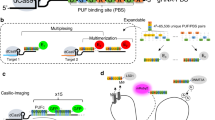
Abstract
Imaging and chromatin capture techniques have provided important insights into our understanding of nuclear organization. A limitation of these techniques is the inability to resolve allele-specific spatiotemporal properties of genomic loci in living cells. Here, we describe an allele-specific CRISPR live-cell DNA imaging technique (SNP-CLING) to provide the first comprehensive insights into allelic positioning across space and time in mouse embryonic stem cells and fibroblasts. With 3D imaging, we studied alleles on different chromosomes in relation to one another and relative to nuclear substructures such as the nucleolus. We find that alleles maintain similar positions relative to each other and the nucleolus; however, loci occupy unique positions. To monitor spatiotemporal dynamics by SNP-CLING, we performed 4D imaging and determined that alleles are either stably positioned or fluctuating during cell state transitions, such as apoptosis. SNP-CLING is a universally applicable technique that enables the dissection of allele-specific spatiotemporal genome organization in live cells.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Dekker, J. Two ways to fold the genome during the cell cycle: insights obtained with chromosome conformation capture. Epigenetics Chromatin 7, 25 (2014).
Lupiáñez, D. G. et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161, 1012–1025 (2015).
Dixon, J. R. et al. Chromatin architecture reorganization during stem cell differentiation. Nature 518, 331–336 (2015).
Gorkin, D. U., Leung, D. & Ren, B. The 3D genome in transcriptional regulation and pluripotency. Cell Stem Cell 14, 762–775 (2014).
Jost, K. L. et al. Gene repositioning within the cell nucleus is not random and is determined by its genomic neighborhood. Epigenetics Chromatin 8, 36 (2015).
Chubb, J. R., Boyle, S., Perry, P. & Bickmore, W. A. Chromatin motion is constrained by association with nuclear compartments in human cells. Curr. Biol. 12, 439–445 (2002).
Kumaran, R. I. & Spector, D. L. A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. J. Cell Biol. 180, 51–65 (2008).
Meaburn, K. J., Gudla, P. R., Khan, S., Lockett, S. J. & Misteli, T. Disease-specific gene repositioning in breast cancer. J. Cell Biol. 187, 801–812 (2009).
Ferrai, C., de Castro, I. J., Lavitas, L., Chotalia, M. & Pombo, A. Gene positioning. Cold Spring Harb. Perspect. Biol. 2, a000588 (2010).
Franke, M. et al. Formation of new chromatin domains determines pathogenicity of genomic duplications. Nature 538, 265–269 (2016).
Weise, A. et al. POD-FISH: a new technique for parental origin determination based on copy number variation polymorphism. Methods Mol. Biol. 659, 291–298 (2010).
Beliveau, B. J. et al. Single-molecule super-resolution imaging of chromosomes and in situ haplotype visualization using Oligopaint FISH probes. Nat. Commun. 6, 7147 (2015).
Giorgetti, L. et al. Predictive polymer modeling reveals coupled fluctuations in chromosome conformation and transcription. Cell 157, 950–963 (2014).
Darrow, E. M. et al. Deletion of DXZ4 on the human inactive X chromosome alters higher-order genome architecture. Proc. Natl. Acad. Sci. USA 113, E4504–E4512 (2016).
Hoffman, E. A., Frey, B. L., Smith, L. M. & Auble, D. T. Formaldehyde crosslinking: a tool for the study of chromatin complexes. J. Biol. Chem. 290, 26404–26411 (2015).
Hacisuleyman, E. et al. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat. Struct. Mol. Biol. 21, 198–206 (2014).
Abe, Y. et al. Xq26.1-26.2 gain identified on array comparative genomic hybridization in bilateral periventricular nodular heterotopia with overlying polymicrogyria. Dev. Med. Child Neurol. 56, 1221–1224 (2014).
Maass, P. G. et al. A misplaced lncRNA causes brachydactyly in humans. J. Clin. Invest. 122, 3990–4002 (2012).
Shechner, D. M., Hacisuleyman, E., Younger, S. T. & Rinn, J. L. Multiplexable, locus-specific targeting of long RNAs with CRISPR-Display. Nat. Methods 12, 664–670 (2015).
Chao, J. A., Patskovsky, Y., Almo, S. C. & Singer, R. H. Structural basis for the coevolution of a viral RNA-protein complex. Nat. Struct. Mol. Biol. 15, 103–105 (2008).
Shcherbakova, D. M. & Verkhusha, V. V. Near-infrared fluorescent proteins for multicolor in vivo imaging. Nat. Methods 10, 751–754 (2013).
Filipovska, A., Razif, M. F., Nygård, K. K. & Rackham, O. A universal code for RNA recognition by PUF proteins. Nat. Chem. Biol. 7, 425–427 (2011).
Cheong, C. G. & Hall, T. M. Engineering RNA sequence specificity of Pumilio repeats. Proc. Natl. Acad. Sci. USA 103, 13635–13639 (2006).
Anders, C., Niewoehner, O., Duerst, A. & Jinek, M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 513, 569–573 (2014).
Doench, J. G. et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34, 184–191 (2016).
Bodnar, A. G. et al. Extension of life-span by introduction of telomerase into normal human cells. Science 279, 349–352 (1998).
Goldring, M. B. et al. Interleukin-1 beta-modulated gene expression in immortalized human chondrocytes. J. Clin. Invest. 94, 2307–2316 (1994).
Nora, E. P. et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485, 381–385 (2012).
Sehgal, N. et al. Large-scale probabilistic 3D organization of human chromosome territories. Hum. Mol. Genet. 25, 419–436 (2016).
Cremer, T. & Cremer, M. Chromosome territories. Cold Spring Harb. Perspect. Biol. 2, a003889 (2010).
Williams, S. R. et al. Haploinsufficiency of HDAC4 causes brachydactyly mental retardation syndrome, with brachydactyly type E, developmental delays, and behavioral problems. Am. J. Hum. Genet. 87, 219–228 (2010).
Beagrie, R. A. et al. Complex multi-enhancer contacts captured by genome architecture mapping. Nature 543, 519–524 (2017).
Shopland, L. S., Johnson, C. V., Byron, M., McNeil, J. & Lawrence, J. B. Clustering of multiple specific genes and gene-rich R-bands around SC-35 domains: evidence for local euchromatic neighborhoods. J. Cell Biol. 162, 981–990 (2003).
Edelman, L. B. & Fraser, P. Transcription factories: genetic programming in three dimensions. Curr. Opin. Genet. Dev. 22, 110–114 (2012).
Lamond, A. I. & Spector, D. L. Nuclear speckles: a model for nuclear organelles. Nat. Rev. Mol. Cell Biol. 4, 605–612 (2003).
Shea, J. R. Jr & Leblond, C. P. Number of nucleoli in various cell types of the mouse. J. Morphol. 119, 425–433 (1966).
Yang, F. et al. The lncRNA Firre anchors the inactive X chromosome to the nucleolus by binding CTCF and maintains H3K27me3 methylation. Genome Biol. 16, 52 (2015).
Kulkarni, G. V. & McCulloch, C. A. Serum deprivation induces apoptotic cell death in a subset of Balb/c 3T3 fibroblasts. J. Cell Sci. 107, 1169–1179 (1994).
Morimoto, M. & Boerkoel, C. F. The role of nuclear bodies in gene expression and disease. Biology (Basel) 2, 976–1033 (2013).
Thul, P. J. et al. A subcellular map of the human proteome. Science 356, eaal3321 (2017).
Chess, A. Monoallelic gene expression in mammals. Annu. Rev. Genet. 50, 317–327 (2016).
Cui, C., Shu, W. & Li, P. Fluorescence in situ hybridization: cell-based genetic diagnostic and research applications. Front. Cell Dev. Biol. 4, 89 (2016).
Wijchers, P. J. et al. Cause and consequence of tethering a SubTAD to different nuclear compartments. Mol. Cell 61, 461–473 (2016).
Peric-Hupkes, D. et al. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol. Cell 38, 603–613 (2010).
Lupiáñez, D. G., Spielmann, M. & Mundlos, S. Breaking TADs: how alterations of chromatin domains result in disease. Trends Genet. 32, 225–237 (2016).
Wijchers, P. J. & de Laat, W. Genome organization influences partner selection for chromosomal rearrangements. Trends Genet. 27, 63–71 (2011).
Doench, J. G. et al. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat. Biotechnol. 32, 1262–1267 (2014).
Engreitz, J. M. et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science 341, 1237973 (2013).
Acknowledgements
We thank all of the Rinn laboratory members for thoughtful discussions and intellectual input. The study was supported by U01 DA040612-01, P01 GM09911, HHMI Faculty Scholars Program (J.L.R), and the ‘Deutsche Forschungsgemeinschaft (DFG)’, supported P.G.M (MA5028/1-3 and MA5028/1-1). M. B. Goldring (Weill Cornell Medical College) kindly provided C28/I2 cells. We thank S. Terclavers (Zeiss) and D. Richardson (Harvard Center for Biological Imaging, HCBI, Cambridge) for their support optimizing Airyscan microscopy, J. P. Lewandowski (Harvard University) for generating 129S1/CAST MEFs, and J. Engreitz (Broad Institute) for providing 129S1/CAST mESCs.
Author information
Authors and Affiliations
Contributions
P.G.M. and J.L.R. conceived the study. P.G.M. performed the experiments and wrote the manuscript with J.L.R and A.R.B. C.L.W. performed the mESC experiments, M.M. analyzed the availability of suitable SNP-CLING SNPs, and D.M.S. provided intellectual input and plasmids.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Genotypization of SNP-CLING sgRNAs.
(a) Genome-wide distribution of suitable SNPs for SNP-CLING in human (mean distance = 181 bp, hg38), and in mouse (mean distance 332 bp, mm10, medians with 25th to 75th percentiles, 1.5x interquartile ranges, outliers = black dots). (b) Total numbers of suitable SNPs in human and mouse, and intergenic / intragenic distributions. (c) Heterozygous SNP substitutions in the second or third nucleotide of the PAM-sequences determined the usability of every sgRNA in SNP-CLING experiments on 129S1/CAST cells. For each allele, two sgRNAs specifically labelled every locus.
Supplementary Figure 2 Confirmation of live-cell imaging and CLING’s specificity.
(a) Very low laser usage implicates less bleaching and phototoxic effects, ensuring the viability of the cells during time-lapse imaging. Thus, we used a power meter to convert linearly the laser output that is set in percent units in the operating ZEN black software (Zeiss) to µWatt (µW). The grey regions in the plots indicate the bandwidths that were used in all SNP-CLING or CLING experiments. Since a cell has a self-power of ~7 µW, the used laser power was suitable for live cell imaging. The laser at 405 nm was used to image Hoechst 33342 9 (n = 1, Pearson’s correlation). (b) 458 nm were used to visualize GFP fluorescence of the nucleolus (n = 1) and (c) 514 nm for mVenus (n = 1, Pearson’s correlation). (d) mCherry was excited with a 594 nm laser (n = 1, Pearson’s correlation). (e) CLING-signals were characterized in detail to get an impression of the variety of accumulated foci in the nucleus, as well as in the cytosol, in the presence or absence of sgRNAs or dCas9. A sgRNA recognizing telomere sequences was used to establish the transient transfection conditions (n = 10), (Chen, Gilbert et al. 2013). (f) Cytosolic signals in addition to nuclear foci occurred very rarely in some cells, independent of the used sgRNA pool (2/10 nuclei, n = 3, arrowheads = CLING signals of FIRRE and non-specific signals). (g) Transfections without the sgRNA pool generated hazy and cloudy signals throughout the nucleus. Accumulated nuclear punctae were not found (n = 10). (h) In very rare cases one or two foci were observed in a transfection without dCas9 (0.5/10 nuclei, n = 4, arrowheads = CLING signals of CISTR-ACT). (i) An alternating order of MS2-PP7-MS2-PP7 stemloops was cloned into a plasmid to determine whether or not the fluorochromes generated signals that overlap. We used the telomeres sgRNA and determined that the fluorochromes bound specifically. At every foci, both fluorochromes were detected as totally overlapped and mixed signals, as it was previously shown for other fluorochromes and repetitive sequences (Ma, Tu et al. 2016).
Supplementary Figure 3 Signal processing and CLING’s specificity.
Signal processing of all acquired SNP-CLING images in (a) mESCs cells, in (b) mEFs, and CLING images in (c) RPE-1 cells. Arrowheads depict specific and non-specific CLING foci. The images collected on the LSM880+Airyscan have increased sensitivity (4-8 x) due to the 32 channel gallium arsenide phosphide photomultiplier tube (GaAsP-PMT) area detector that collects a pinhole-plane image at every scan position (Huff 2015). This increased sensitivity results in detection of both specific and background fluorochrome-derived signals in each image, which can be differentiated in post-acquisition image processing (described below and in methods). The number, sizes and brightness of non-specific background signals vary depending on the expression of the transfected fluorescent proteins and the imaging conditions (laser power, see Figure S2). The number of visible signals corresponding with the known number of alleles in a given cell line were obvious in the live view. For post-acquisition image analysis, z-stack planes were first merged by performing a maximum intensity projection (MIP). The ‘best fit’ analysis option was used to preliminarily adjust intensity thresholds. X-Y dimensions at this stage are shown in the top row. In sequential manual adjustment steps, signal from background haze, random fluorescent protein accumulation, and/or potential off-target dCas9 binding events were removed (second and third row). Signals present in final processed images (bottom row) correspond to expected karyotypes: (a) a single signal for the sole Firre locus and a single signal for the allele-specifically labeled Ypel4 locus were detected in male mESCs; (b) a single signal for the allele-specifically labeled Firre locus was detected in female mEFs; (c) two signals for either the CISTR-ACT or the FIRRE loci were detected in diploid RPE-1 cells (Darrow, Huntley et al. 2016). To investigate the difference in signal intensities between specific and non-specific signals in all cell types, we calculated the relative signal intensities by generating the median intensity of all detected signals in a given nucleus. The median intensity was used to normalize all detected signals. The intensities as ratios are represented as bar graphs below the corresponding image and show that the specific signals were the brightest. Of note, specific signal intensities were at least two-fold higher than non-specific signals. In validation experiments without the transfection of sgRNAs, no bright nuclear foci were detected (see Figure S2g). (d, e) We also measured the three-dimensional sizes (x, y, z dimensions) of all foci detected by the ‘best fit’ analysis. The signals that corresponded with processed specific signals were the biggest signals, as can be seen in represented image panel (d). We further quantified the difference in signal size with measurements from at least 30 nuclei (e). The specific ‘biggest’ signals were present in 12.7±1.98 z-stack planes (each 0.17 µm), in comparison to the smaller background signals that were detected in 5.3±1.5 planes. In all instances, the brightest and biggest signals correspond to the known number of alleles in RPE-1, mESCs and mEFs. (f) To further address CLING’s specificity, we quantified signals of CISTR-ACT, XIST, and SOX9 in human RPE-1 cells in the presence and absence of dCas9. As expected, dCas9’s presence dramatically increased the occurrence of specific signals. On average, 30 % of transfected cells showed the expected diploid status (two signals), whilst some cells showed only one signal, and ˜30 % of transfected cells showed either no or many signals. (g) In SNP-CLING, we determined a specific separation between the parental alleles in 83 % of the imaged nuclei in hybrid 129S1/CAST MEFs. The remaining 17 % of cells showed either no or multiple signals of the second allele, or cross-labeling of one of the two alleles. Of the 83 % of cells, 12 % showed two foci, indicating mitotic stage G2. (h) Haploid signals of sgRNAs targeting either the 129S1 (75 %) or CAST (90 %) allele determined SNP-CLING’s specificity in hybrid 129S1/CAST MEFs. (i) The co-localization frequency (78 %) of targeting CISTR-ACT with orthologous MS2 and PP7 sgRNA pools was highly specific in RPE-1 cells. (j) In ˜6 of 10 cells, a clear separation between XIST and TSIX was achieved (no co-localization) in RPE-1 cells. The heterochromatin formation of the inactive X chromosome and different chromatin compaction rates may also influence the spatial distances between two given loci.
Supplementary Figure 4 SNP-CLING signals of mouse loci.
(a) Gene-density (Gencode vM9) and chromosomal sizes (mm10) of the mouse genome. Loci on chromosome 1 (large chromosome), chromosome 7 and 11 (gene-rich chromosomes), chromosome 15 (gene-poor / small chromosome), and chromosome 18 (gene-poor / small chromosome), and Firre on chromosome X, were selected for SNP-CLING experiments. (b) Examples of allele-specific SNP-CLING signals (separate channels) of maternal and paternal loci in combination with rRNA staining of the nucleoli in female 129S1/CAST mEFs (n = 10, arrowheads = specific SNP-CLING foci of maternal and paternal alleles, or CLING foci for chr. 18 Puf-Pum1-iRFP670).
Supplementary Figure 5 CLING signals of mouse and human loci.
Examples of labeling various human and mouse loci with sgRNA pools. Four different sgRNAs were mixed in combinations of three to determine the best set for imaging (arrowheads = specific CLING foci). Before relevant experiments, the use of each sgRNA pool was specifically tested for (a) mouse loci in male mESCs (Firre on chromosome X was therefore detected as single voxel, n = 10), and (b) for coding genes in human in human RPE-1 or C28/I2 cells (n = 10).
Supplementary Figure 6 Data points of individual samples of figure 5.
(a) Individual data points of figure 5b, (b) = figure 5c, (c) = figure 5d, (d) = figure 5h, (e) = figure 5i, (f) = figure 5k, (g) = figure 5l.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 and Supplementary Tables 1 and 2
Rights and permissions
About this article
Cite this article
Maass, P.G., Barutcu, A.R., Shechner, D.M. et al. Spatiotemporal allele organization by allele-specific CRISPR live-cell imaging (SNP-CLING). Nat Struct Mol Biol 25, 176–184 (2018). https://doi.org/10.1038/s41594-017-0015-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-017-0015-3
This article is cited by
-
DNA double-strand break–capturing nuclear envelope tubules drive DNA repair
Nature Structural & Molecular Biology (2024)
-
Live-cell imaging of chromatin contacts opens a new window into chromatin dynamics
Epigenetics & Chromatin (2023)
-
CRISPR imaging reveals chromatin fluctuation at the centromere region related to cellular senescence
Scientific Reports (2023)
-
CRISPR-mediated multiplexed live cell imaging of nonrepetitive genomic loci with one guide RNA per locus
Nature Communications (2022)
-
Recent advances of the biological and biomedical applications of CRISPR/Cas systems
Molecular Biology Reports (2022)