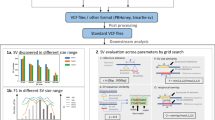
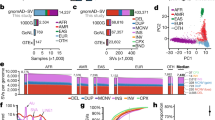
Abstract
The availability of long reads is revolutionizing studies of structural variants (SVs). However, because SVs vary across individuals and are discovered through imprecise read technologies and methods, they can be difficult to compare. Addressing this, we present Jasmine and Iris (https://github.com/mkirsche/Jasmine/), for fast and accurate SV refinement, comparison and population analysis. Using an SV proximity graph, Jasmine outperforms six widely used comparison methods, including reducing the rate of Mendelian discordance in trio datasets by more than fivefold, and reveals a set of high-confidence de novo SVs confirmed by multiple technologies. We also present a unified callset of 122,813 SVs and 82,379 indels from 31 samples of diverse ancestry sequenced with long reads. We genotype these variants in 1,317 samples from the 1000 Genomes Project and the Genotype-Tissue Expression project with DNA and RNA-sequencing data and assess their widespread impact on gene expression, including within medically relevant genes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The sequencing data used in this study are available from the publications listed in Supplementary Table 1 and Supplementary Table 2. All variant calls and associations are available at http://data.schatz-lab.org/jasmine/.
Code availability
The Jasmine and Iris code and documentation are available open source at https://github.com/mkirsche/Jasmine/ and https://github.com/mkirsche/Iris/. The versions used in the paper are archived in Zenodo for Jasmine62 and Iris63. These methods are also available in Bioconda and Galaxy to simplify use on the command line or within the Galaxy graphical user interface. The versions of all software packages used in the manuscript are described in Supplementary Table 3.
References
Alonge, M. et al. Major impacts of widespread structural variation on gene expression and crop improvement in tomato. Cell 182, 145–161 (2020).
Alkan, C., Coe, B. P. & Eichler, E. E. Genome structural variation discovery and genotyping. Nat. Rev. Genet. 12, 363–376 (2011).
Chiang, C. et al. The impact of structural variation on human gene expression. Nat. Genet. 49, 692–699 (2017).
Aganezov, S. et al. Comprehensive analysis of structural variants in breast cancer genomes using single-molecule sequencing. Genome Res. 30, 1258–1273 (2020).
Nattestad, M. et al. Complex rearrangements and oncogene amplifications revealed by long-read DNA and RNA sequencing of a breast cancer cell line. Genome Res. 28, 1126–1135 (2018).
Brandler, W. M. et al. Paternally inherited cis-regulatory structural variants are associated with autism. Science 360, 327–331 (2018).
Jeffares, D. C. et al. Transient structural variations have strong effects on quantitative traits and reproductive isolation in fission yeast. Nat. Commun. 8, 14061 (2017).
Sedlazeck, F. J., Lee, H., Darby, C. A. & Schatz, M. C. Piercing the dark matter: bioinformatics of long-range sequencing and mapping. Nat. Rev. Genet. 19, 329–346 (2018).
Mahmoud, M. et al. Structural variant calling: the long and the short of it. Genome Biol. 20, 246 (2019).
Zook, J. M. et al. Integrating human sequence data sets provides a resource of benchmark SNP and indel genotype calls. Nat. Biotechnol. 32, 246–251 (2014).
Sirén, J. et al. Pangenomics enables genotyping of known structural variants in 5202 diverse genomes. Science 374, abg8871 (2021).
Narzisi, G. et al. Accurate de novo and transmitted indel detection in exome-capture data using microassembly. Nat. Methods 11, 1033–1036 (2014).
Korlach, J. et al. Real-time DNA sequencing from single polymerase molecules. Methods Enzymol. 472, 431–455 (2010).
Jain, M., Olsen, H. E., Paten, B. & Akeson, M. The Oxford Nanopore MinION: delivery of nanopore sequencing to the genomics community. Genome Biol. 17, 239 (2016).
Wenger, A. M. et al. Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat. Biotechnol. 37, 1155–1162 (2019).
Goodwin, S., McPherson, J. D. & McCombie, W. R. Coming of age: ten years of next-generation sequencing technologies. Nat. Rev. Genet. 17, 333–351 (2016).
Jain, C., Rhie, A., Hansen, N. F., Koren, S. & Phillippy, A. M. Long-read mapping to repetitive reference sequences using Winnowmap2. Nat. Methods https://doi.org/10.1038/s41592-022-01457-8 (2022).
Sedlazeck, F. J. et al. Accurate detection of complex structural variations using single-molecule sequencing. Nat. Methods 15, 461–468 (2018).
Li, H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018).
Jiang, T. et al. Long-read-based human genomic structural variation detection with cuteSV. Genome Biol. 21, 189 (2020).
Chaisson, M. J. P. et al. Multi-platform discovery of haplotype-resolved structural variation in human genomes. Nat. Commun. 10, 1784 (2019).
Audano, P. A. et al. Characterizing the major structural variant alleles of the human genome. Cell 176, 663–675 (2019).
Beyter, D. et al. Long-read sequencing of 3,622 Icelanders provides insight into the role of structural variants in human diseases and other traits. Nat. Genet. https://doi.org/10.1038/s41588-021-00865-4 (2021).
Byrska-Bishop, M. et al. High-coverage whole-genome sequencing of the expanded 1000 Genomes Project cohort including 602 trios. Cell 185, 3426–3440 (2022).
GTEx Consortium. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 369, 1318–1330 (2020).
Vaser, R., Sović, I., Nagarajan, N. & Šikić, M. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 27, 737–746 (2017).
Kruskal, J. B. On the shortest spanning subtree of a graph and the traveling salesman problem. Proc. Am. Math. Soc. https://doi.org/10.1090/s0002-9939-1956-0078686-7 (1956).
Bentley, J. L. Multidimensional binary search trees used for associative searching. Comm. ACM https://doi.org/10.1145/361002.361007 (1975).
Jalili, V. et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2020 update. Nucleic Acids Res. 48, W395–W402 (2020).
Iossifov, I. et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature 515, 216–221 (2014).
Renaux-Petel, M. et al. Contribution of de novo and mosaic mutations to Li-Fraumeni syndrome. J. Med. Genet. 55, 173–180 (2018).
Veltman, J. A. & Brunner, H. G. De novo mutations in human genetic disease. Nat. Rev. Genet. https://doi.org/10.1038/nrg3241 (2012).
Belyeu, J. R. et al. De novo structural mutation rates and gamete-of-origin biases revealed through genome sequencing of 2,396 families. Am. J. Hum. Genet. 108, 597–607 (2021).
Shi, J. et al. Structural variant selection for high-altitude adaptation using single-molecule long-read sequencing. Preprint at bioRxiv https://doi.org/10.1101/2021.03.27.436702 (2021).
Ebert, P. et al. Haplotype-resolved diverse human genomes and integrated analysis of structural variation. Science 372, eabf7117 (2021).
Larson, D. E. et al. svtools: population-scale analysis of structural variation. Bioinformatics 35, 4782–4787 (2019).
Eggertsson, H. P. et al. GraphTyper2 enables population-scale genotyping of structural variation using pangenome graphs. Nat. Commun. 10, 5402 (2019).
Cooper, G. M. et al. A copy number variation morbidity map of developmental delay. Nat. Genet. 43, 838–846 (2011).
Zook, J. M. et al. A robust benchmark for detection of germline large deletions and insertions. Nat. Biotechnol. 38, 1347–1355 (2020).
Ellegren, H. Microsatellites: simple sequences with complex evolution. Nat. Rev. Genet. https://doi.org/10.1038/nrg1348 (2004).
Ranallo-Benavidez, T. R. et al. Optimized sample selection for cost-efficient long-read population sequencing. Genome Res. https://doi.org/10.1101/gr.264879.120 (2021).
Consortium, T. 1000 G. P. & The 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature https://doi.org/10.1038/nature15393 (2015).
Chen, S. et al. Paragraph: a graph-based structural variant genotyper for short-read sequence data. Genome Biol. 20, 291 (2019).
Lappalainen, T. et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature 501, 506–511 (2013).
Hormozdiari, F., Kostem, E., Kang, E. Y., Pasaniuc, B. & Eskin, E. Identifying causal variants at loci with multiple signals of association. Genetics 198, 497–508 (2014).
Schatz, M. C. et al. Inverting the model of genomics data sharing with the NHGRI Genomic Data Science Analysis, Visualization, and Informatics Lab-space. Cell Genom. 2, 100085 (2022).
Scott, A. J., Chiang, C. & Hall, I. M. Structural variants are a major source of gene expression differences in humans and often affect multiple nearby genes. Genome Res. https://doi.org/10.1101/gr.275488.121 (2021).
Mezzar, S. et al. Phytol-induced pathology in 2-hydroxyacyl-CoA lyase (HACL1) deficient mice. Evidence for a second non-HACL1-related lyase. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1862, 972–990 (2017).
Caltabiano, R. et al. Macrophage migration inhibitory factor (MIF) and its homologue d-dopachrome tautomerase (DDT) inversely correlate with inflammation in discoid lupus erythematosus. Molecules 26, 184 (2021).
Torres-Mora, J. et al. Malignant melanotic schwannian tumor: a clinicopathologic, immunohistochemical, and gene expression profiling study of 40 cases, with a proposal for the reclassification of ‘melanotic schwannoma’. Am. J. Surg. Pathol. 38, 94–105 (2014).
Nurk, S. et al. The complete sequence of a human genome. Science 376, 44–53 (2022).
Aganezov, S. et al. A complete reference genome improves analysis of human genetic variation. Science 376, eabl3533 (2022).
Wigginton, J. E., Cutler, D. J. & Abecasis, G. R. A note on exact tests of Hardy–Weinberg equilibrium. Am. J. Hum. Genet. 76, 887–893 (2005).
Navarro Gonzalez, J. et al. The UCSC Genome Browser database: 2021 update. Nucleic Acids Res. 49, D1046–D1057 (2021).
Zerbino, D. R., Wilder, S. P., Johnson, N., Juettemann, T. & Flicek, P. R. The Ensembl regulatory build. Genome Biol. 16, 56 (2015).
Fu, Y. et al. FunSeq2: a framework for prioritizing noncoding regulatory variants in cancer. Genome Biol. 15, 480 (2014).
Abel, H. J. et al. Mapping and characterization of structural variation in 17,795 human genomes. Nature 583, 83–89 (2020).
Rentzsch, P., Witten, D., Cooper, G. M., Shendure, J. & Kircher, M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 47, D886–D894 (2019).
Huang, Y.-F., Gulko, B. & Siepel, A. Fast, scalable prediction of deleterious noncoding variants from functional and population genomic data. Nat. Genet. 49, 618–624 (2017).
Hubisz, M. J., Pollard, K. S. & Siepel, A. PHAST and RPHAST: phylogenetic analysis with space/time models. Brief. Bioinform. 12, 41–51 (2011).
Chuang, L.-S. et al. A frameshift in CSF2RB predominant among Ashkenazi Jews increases risk for Crohn’s disease and reduces monocyte signaling via GMCSF. Gastroenterology 151, 710–723 (2016).
Kirsche, M. Jasmine: Population-scale structural variant merging. Jasmine software release v1.1.0 from https://github.com/mkirsche/Jasmine. Zenodo. https://doi.org/10.5281/zenodo.5586905 (2021).
Kirsche, M. Iris: Structural variant breakpoint and sequence refinement. Iris software release v1.0.4 from https://github.com/mkirsche/Iris. Zenodo. https://doi.org/10.5281/zenodo.5586965 (2021).
Acknowledgements
We thank F. Sedlazeck and M. Alonge for helpful discussions. This work was supported, in part, by National Science Foundation grants DBI-1350041 (to M.C.S.), IOS-1732253 (to M.C.S.) and IOS-1758800 (to M.C.S.) and National Institutes of Health grants NCI U01CA253481 (to M.C.S.), NCI U24CA231877 (to M.C.S.), NHGRI U41HG006620 (to M.C.S.), NHGRI U24HG010263 (to M.C.S.), NIH R03CA272952 (to M.C.S.) and NIGMS R35GM139580 (to A.B.). This work was also supported in part by the Mark Foundation for Cancer Research award 19-033-ASP (to M.C.S.) and a Microsoft Research Fellows award (to A.B.). Part of this research project was conducted using computational resources at the Maryland Advanced Research Computing Center (MARCC). We also thank the investigators and the patient donors from the Human Pangenome Reference Consortium, GTEx, and 1000 Genomes for making their data available.
Author information
Authors and Affiliations
Contributions
M.K. was the principal author of the Jasmine and Iris software, and led most of the presented analyses. G.P. contributed to the genotyping and eQTL analysis of the 1000 Genomes cohort. R.S. contributed to the genotyping of the 1000 Genomes cohort. B.N. led the genotyping and eQTL analysis of the GTEx cohort. A.B. assisted in the analysis of the GTEx cohort. S.A. helped design the software methods and the overall research strategy. M.C.S. oversaw all aspects of the research and analysis. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
S.A. has become an employee at Oxford Nanopore. R.S. has become an employee at Illumina. M.K. has become an employee at Variant Bio.
Peer review
Peer review information
Nature Methods thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editor: Lin Tang, in collaboration with the Nature Methods team. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Tables 1–3, Figs. 1–47 and Notes 1 and 2.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kirsche, M., Prabhu, G., Sherman, R. et al. Jasmine and Iris: population-scale structural variant comparison and analysis. Nat Methods 20, 408–417 (2023). https://doi.org/10.1038/s41592-022-01753-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-022-01753-3
This article is cited by
-
De novo and somatic structural variant discovery with SVision-pro
Nature Biotechnology (2024)
-
Pangenome graphs improve the analysis of structural variants in rare genetic diseases
Nature Communications (2024)
-
Diallel panel reveals a significant impact of low-frequency genetic variants on gene expression variation in yeast
Molecular Systems Biology (2024)
-
A sequence-aware merger of genomic structural variations at population scale
Nature Communications (2024)
-
Plant pangenomes for crop improvement, biodiversity and evolution
Nature Reviews Genetics (2024)