Abstract
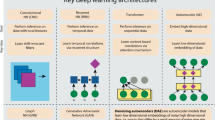
To enable the application of deep learning in biology, we present Selene (https://selene.flatironinstitute.org/), a PyTorch-based deep learning library for fast and easy development, training, and application of deep learning model architectures for any biological sequence data. We demonstrate on DNA sequences how Selene allows researchers to easily train a published architecture on new data, develop and evaluate a new architecture, and use a trained model to answer biological questions of interest.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Code availability
Selene is open-source software (license BSD 3-Clause Clear). Project homepage: https://selene.flatironinstitute.org. GitHub: https://github.com/FunctionLab/selene. Archived version: https://github.com/FunctionLab/selene/archive/0.2.0.tar.gz.
Data availability
Cistrome14, Cistrome file ID 33545, measurements from GSM970258: http://dc2.cistrome.org/api/downloads/eyJpZCI6IjMzNTQ1In0%3A1fujCu%3ArNvWLCNoET6o9SdkL8fEv13uRu4b/. ENCODE21 and Roadmap Epigenomics22 chromatin profiles: files listed in Supplementary Table 1 of ref. 4. IGAP age at onset survival16,17: https://www.niagads.org/datasets/ng00058 (P-values-only file). The case studies used processed datasets from these sources. They can be downloaded at the following Zenodo links: Cistrome, https://zenodo.org/record/2214130/files/data.tar.gz; ENCODE and Roadmap Epigenomics chromatin profiles, https://zenodo.org/record/2214970/files/chromatin_profiles.tar.gz; IGAP age at onset survival, https://zenodo.org/record/1445556/files/variant_effect_prediction_data.tar.gz. Source data for Figs. 2 and 3 are available online.
References
LeCun, Y., Bengio, Y. & Hinton, G. Nature 521, 436–444 (2015).
Ching, T. et al. J. R. Soc. Interface. 15, 20170387 (2018).
Segler, M. H. S., Preuss, M. & Waller, M. P. Nature 555, 604–610 (2018).
Zhou, J. & Troyanskaya, O. G. Nat. Meth. 12, 931–934 (2015).
Alipanahi, B., Delong, A., Weirauch, M. T. & Frey, B. J. Nat. Biotechnol. 33, 831–838 (2015).
Kelley, D. R., Snoek, J. & Rinn, J. L. Genome Res. 26, 990–999 (2016).
Angermueller, C., Lee, H. J., Reik, W. & Stegle, O. Genome. Biol. 18, 67 (2017).
Kelley, D. R. et al. Genome Res. 28, 739–750 (2018).
Quang, D. & Xie, X. Nucleic Acids Res. 44, e107 (2016).
Sundaram, L. et al. Nat. Genet. 50, 1161–1170 (2018).
Min, S., Lee, B. & Yoon, S. Brief. Bioinform. 18, 851–869 (2017).
Budach, S. & Marsico, A. Bioinformatics 34, 3035–3037 (2018).
Avsec, Z. et al. bioRxiv Preprint at https://www.biorxiv.org/content/10.1101/375345v1 (2018).
Mei, S. et al. Nucleic Acids Res. 45, D658–D662 (2017).
Troyanskaya, O. G. et al. Selene CLI operations and outputs. Selene https://selene.flatironinstitute.org/overview/cli.html (2018).
Ruiz, A. et al. Transl. Psychiatry 4, e358 (2014).
Huang, K.-L. et al. Nat. Neurosci. 20, 1052–1061 (2017).
Li, H. et al. Bioinformatics 25, 2078–2079 (2009).
Li, H. Bioinformatics 27, 718–719 (2011).
ENCODE Project. Reference sequences. ENCODE: Encyclopedia of DNA Elements https://www.encodeproject.org/data-standards/reference-sequences/ (2016).
ENCODE Project Consortium. Nature 489, 57–74 (2012).
Kundaje, A. et al. Nature 518, 317–330 (2015).
Acknowledgements
The authors acknowledge all members of the Troyanskaya lab for helpful discussions. In addition, the authors thank D. Simon for setting up the website and automating updates to the site. The authors are pleased to acknowledge that this work was performed using the high-performance computing resources at Simons Foundation and the TIGRESS computer center at Princeton University. This work was supported by NIH grants R01HG005998, U54HL117798, R01GM071966, and T32HG003284; HHS grant HHSN272201000054C; and Simons Foundation grant 395506, all to O.G.T. O.G.T. is a CIFAR fellow.
Author information
Authors and Affiliations
Contributions
K.M.C and J.Z. conceived the Selene library. K.M.C. and E.M.C. designed, implemented, and documented Selene. K.M.C. performed the analyses described in the manuscript. O.G.T. supervised the project. K.M.C., E.M.C., and O.G.T wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
About this article
Cite this article
Chen, K.M., Cofer, E.M., Zhou, J. et al. Selene: a PyTorch-based deep learning library for sequence data. Nat Methods 16, 315–318 (2019). https://doi.org/10.1038/s41592-019-0360-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-019-0360-8
This article is cited by
-
Fundamentals for predicting transcriptional regulations from DNA sequence patterns
Journal of Human Genetics (2024)
-
Predicting the impact of sequence motifs on gene regulation using single-cell data
Genome Biology (2023)
-
Predictive analyses of regulatory sequences with EUGENe
Nature Computational Science (2023)
-
From macro to micro: rethinking multi-scale pedestrian detection
Multimedia Systems (2023)
-
Histone-Net: a multi-paradigm computational framework for histone occupancy and modification prediction
Complex & Intelligent Systems (2023)