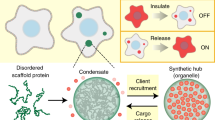
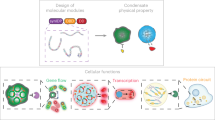
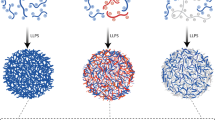
Abstract
Protein condensates are distinct structures assembled in living cells that concentrate molecules via phase separation in a confined subcellular compartment. In the past decade, remarkable advances have been made to discover the fundamental roles of the condensates in spatiotemporal control of cellular metabolism and physiology and to reveal the molecular principles, components and driving forces that underlie their formation. Here we review the unique properties of the condensates, the promise and hurdles for harnessing them toward purposeful design and manipulation of biological functions in living cells. In particular, we highlight recent advances in mining and understanding the proteinaceous components for creating designer condensates, along with the engineering approaches to manipulate their material properties and biological functions. With these advances, a greater variety of complex organelle-like structures can be built for diverse applications, with unprecedented effects on synthetic biology.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Boeynaems, S. et al. Protein phase separation: a new phase in cell biology. Trends Cell Biol. 28, 420–435 (2018).
Feng, Z., Chen, X., Wu, X. & Zhang, M. Formation of biological condensates via phase separation: characteristics, analytical methods, and physiological implications. J. Biol. Chem. 294, 14823–14835 (2019).
Zhang, H. et al. Liquid–liquid phase separation in biology: mechanisms, physiological functions and human diseases. Sci. China Life Sci. 63, 953–985 (2020).
Schuster, B. S. et al. Biomolecular condensates: sequence determinants of phase separation, microstructural organization, enzymatic activity, and material properties. J. Phys. Chem. B 125, 3441–3451 (2021).
Reinkemeier, C. D. & Lemke, E. A. Synthetic biomolecular condensates to engineer eukaryotic cells. Curr. Opin. Chem. Biol. 64, 174–181 (2021).
Meng, F. & Ellis, T. The second decade of synthetic biology: 2010–2020. Nat. Commun. 11, 5174 (2020).
Korkmazhan, E., Tompa, P. & Dunn, A. R. The role of ordered cooperative assembly in biomolecular condensates. Nat. Rev. Mol. Cell Biol. 22, 647–648 (2021).
Shin, Y. & Brangwynne, C. P. Liquid phase condensation in cell physiology and disease. Science 357, eaaf4382 (2017).
Jawerth, L. et al. Protein condensates as aging Maxwell fluids. Science 370, 1317–1323 (2020).
Woodruff, J. B., Hyman, A. A. & Boke, E. Organization and function of non-dynamic biomolecular condensates. Trends Biochem. Sci. 43, 81–94 (2018).
Vendruscolo, M. & Fuxreiter, M. Sequence determinants of the aggregation of proteins within condensates generated by liquid–liquid phase separation. J. Mol. Biol. 434, 167201 (2022).
Parry, BradleyR. et al. The bacterial cytoplasm has glass-like properties and is fluidized by metabolic activity. Cell 156, 183–194 (2014).
Alberti, S. & Hyman, A. A. Biomolecular condensates at the nexus of cellular stress, protein aggregation disease and ageing. Nat. Rev. Mol. Cell Biol. 22, 196–213 (2021).
Wiegand, T. & Hyman, A. A. Drops and fibers—how biomolecular condensates and cytoskeletal filaments influence each other. Emerg. Top. Life Sci. 4, 247–261 (2020).
Wei, S.-P. et al. Formation and functionalization of membraneless compartments in Escherichia coli. Nat. Chem. Biol. 16, 1143–1148 (2020). The first report to create functional membraneless compartments using synthetic disordered proteins in prokaryotic Escherichia coli.
Zhao, E. M. et al. Light-based control of metabolic flux through assembly of synthetic organelles. Nat. Chem. Biol. 15, 589–597 (2019). The first report to redirect metabolic flux of a branch pathway in yeast using light-switchable synthetic organelles.
Hastings, R. L. & Boeynaems, S. Designer condensates: a toolkit for the biomolecular architect. J. Mol. Biol. 433, 166837 (2021).
Wiedner, H. J. & Giudice, J. It’s not just a phase: function and characteristics of RNA-binding proteins in phase separation. Nat. Struct. Mol. Biol. 28, 465–473 (2021).
Azaldegui, C. A., Vecchiarelli, A. G. & Biteen, J. S. The emergence of phase separation as an organizing principle in bacteria. Biophys. J. 120, 1123–1138 (2021).
Heidenreich, M. et al. Designer protein assemblies with tunable phase diagrams in living cells. Nat. Chem. Biol. 16, 939–945 (2020).
Maharana, S. et al. RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science 360, 918–921 (2018).
Conicella, A. E. et al. TDP-43 alpha-helical structure tunes liquid–liquid phase separation and function. Proc. Natl Acad. Sci. USA 117, 5883–5894 (2020).
Elbaum-Garfinkle, S. et al. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc. Natl Acad. Sci. USA 112, 7189–7194 (2015).
Hondele, M. et al. DEAD-box ATPases are global regulators of phase-separated organelles. Nature 573, 144–148 (2019).
Holmes, J. A. et al. Caulobacter PopZ forms an intrinsically disordered hub in organizing bacterial cell poles. Proc. Natl Acad. Sci. USA 113, 12490–12495 (2016).
Lasker, K. et al. Selective sequestration of signalling proteins in a membraneless organelle reinforces the spatial regulation of asymmetry in Caulobacter crescentus. Nat. Microbiol. 5, 418–429 (2020).
Wunder, T. & Mueller-Cajar, O. Biomolecular condensates in photosynthesis and metabolism. Curr. Opin. Plant Biol. 58, 1–7 (2020).
Zang, K., Wang, H., Hartl, F. U. & Hayer-Hartl, M. Scaffolding protein CcmM directs multiprotein phase separation in β-carboxysome biogenesis. Nat. Struct. Mol. Biol. 28, 909–922 (2021).
Reichheld, S. E., Muiznieks, L. D., Keeley, F. W. & Sharpe, S. Direct observation of structure and dynamics during phase separation of an elastomeric protein. Proc. Natl Acad. Sci. USA 114, E4408–E4415 (2017).
Dzuricky, M., Rogers, B. A., Shahid, A., Cremer, P. S. & Chilkoti, A. De novo engineering of intracellular condensates using artificial disordered proteins. Nat. Chem. 12, 814–825 (2020).
Simon, J. R., Carroll, N. J., Rubinstein, M., Chilkoti, A. & Lopez, G. P. Programming molecular self-assembly of intrinsically disordered proteins containing sequences of low complexity. Nat. Chem. 9, 509–515 (2017).
Wang, X. et al. LLPSDB v2.0: an updated database of proteins undergoing liquid–liquid phase separation in vitro. Bioinformatics 38, 2010–2014 (2022).
Meszaros, B. et al. PhaSePro: the database of proteins driving liquid–liquid phase separation. Nucleic Acids Res. 48, D360–D367 (2020).
Vernon, R. M. & Forman-Kay, J. D. First-generation predictors of biological protein phase separation. Curr. Opin. Struct. Biol. 58, 88–96 (2019).
van Mierlo, G. et al. Predicting protein condensate formation using machine learning. Cell Rep. 34, 108705 (2021).
Saar, K. L. et al. Learning the molecular grammar of protein condensates from sequence determinants and embeddings. Proc. Natl Acad. Sci. USA 118, e2019053118 (2021).
Dueber, J. E. et al. Synthetic protein scaffolds provide modular control over metabolic flux. Nat. Biotechnol. 27, 753–759 (2009).
Behrendorff, J., Borras-Gas, G. & Pribil, M. Synthetic protein scaffolding at biological membranes. Trends Biotechnol. 38, 432–446 (2020).
Wang, X., Chen, X. & Yang, Y. Spatiotemporal control of gene expression by a light-switchable transgene system. Nat. Methods 9, 266–269 (2012).
Strickland, D. et al. TULIPs: tunable, light-controlled interacting protein tags for cell biology. Nat. Methods 9, 379–384 (2012).
Kennedy, M. J. et al. Rapid blue-light–mediated induction of protein interactions in living cells. Nat. Methods 7, 973–975 (2010).
Taslimi, A. et al. An optimized optogenetic clustering tool for probing protein interaction and function. Nat. Commun. 5, 4925 (2014).
Redchuk, T. A., Omelina, E. S., Chernov, K. G. & Verkhusha, V. V. Near-infrared optogenetic pair for protein regulation and spectral multiplexing. Nat. Chem. Biol. 13, 633–639 (2017).
Toettcher, J. E., Weiner, O. D. & Lim, W. A. Using optogenetics to interrogate the dynamic control of signal transmission by the Ras/Erk module. Cell 155, 1422–1434 (2013).
Kaberniuk, A. A., Shemetov, A. A. & Verkhusha, V. V. A bacterial phytochrome-based optogenetic system controllable with near-infrared light. Nat. Methods 13, 591–597 (2016).
Piraner, D. I., Abedi, M. H., Moser, B. A., Lee-Gosselin, A. & Shapiro, M. G. Tunable thermal bioswitches for in vivo control of microbial therapeutics. Nat. Chem. Biol. 13, 75–80 (2017).
Hill, Z. B., Martinko, A. J., Nguyen, D. P. & Wells, J. A. Human antibody-based chemically induced dimerizers for cell therapeutic applications. Nat. Chem. Biol. 14, 112–117 (2018).
Piraner, D. I., Wu, Y. & Shapiro, M. G. Modular thermal control of protein dimerization. ACS Synth. Biol. 8, 2256–2262 (2019).
Shin, Y. et al. Spatiotemporal control of intracellular phase transitions using light-activated optoDroplets. Cell 168, 159–171 (2017).
Bracha, D. et al. Mapping local and global liquid phase behavior in living cells using photo-oligomerizable seeds. Cell 175, 1467–1480 (2018). The first report to map full intracellular phase diagrams of IDR-rich proteins in living cells by developing a biomimetic photo-activated Corelets system.
Dine, E., Gil, A. A., Uribe, G., Brangwynne, C. P. & Toettcher, J. E. Protein phase separation provides long-term memory of transient spatial stimuli. Cell Syst. 6, 655–663 (2018).
Hong, K., Song, D. & Jung, Y. Behavior control of membrane-less protein liquid condensates with metal ion-induced phase separation. Nat. Commun. 11, 5554 (2020).
Babinchak, W. M. et al. Small molecules as potent biphasic modulators of protein liquid–liquid phase separation. Nat. Commun. 11, 5574 (2020). The first report to demonstrate that the small molecule bis-ANS and similar compounds can act as potent modulators of liquid protein condensates.
Schuster, B. S. et al. Controllable protein phase separation and modular recruitment to form responsive membraneless organelles. Nat. Commun. 9, 2985 (2018).
Kichuk, T. C., Carrasco-Lopez, C. & Avalos, J. L. Lights up on organelles: optogenetic tools to control subcellular structure and organization. WIREs Mech. Dis. 13, e1500 (2021).
Wang, Z. et al. Material properties of phase-separated TFEB condensates regulate the autophagy-lysosome pathway. J. Cell Biol. 221, e202112024 (2022).
Jonchhe, S., Pan, W., Pokhrel, P. & Mao, H. Small molecules modulate liquid-to-solid transitions in phase-separated Tau condensates. Angew. Chem. Int. Ed. 61, e202113156 (2022).
Kroschwald, S., Maharana, S. & Simon, A. Hexanediol: a chemical probe to investigate the material properties of membrane-less compartments. Matters https://doi.org/10.19185/matters.201702000010 (2017).
Hernandez-Candia, C. N., Pearce, S. & Tucker, C. L. A modular tool to query and inducibly disrupt biomolecular condensates. Nat. Commun. 12, 1809 (2021). The first report to inducibly trigger disassembly of user-specified condensates by recruiting a ligand to the scaffold protein.
Chiesa, G., Kiriakov, S. & Khalil, A. S. Protein assembly systems in natural and synthetic biology. BMC Biol. 18, 35 (2020).
Shin, Y. et al. Liquid nuclear condensates mechanically sense and restructure the genome. Cell 175, 1481–1491 (2018).
Schneider, N. et al. Liquid–liquid phase separation of light-inducible transcription factors increases transcription activation in mammalian cells and mice. Sci. Adv. 7, eabd3568 (2021).
Wu, J. et al. Modulating gene regulation function by chemically controlled transcription factor clustering. Nat. Commun. 13, 2663 (2022).
Lin, D. W. et al. Construction of intracellular asymmetry and asymmetric division in Escherichia coli. Nat. Commun. 12, 888 (2021).
Hong, J. C. et al. Localized proteolysis for the construction of intracellular asymmetry in Escherichia coli. ACS Synth. Biol. 10, 1830–1836 (2021).
Veling, M. T. et al. Natural and designed proteins inspired by extremotolerant organisms can form condensates and attenuate apoptosis in human cells. ACS Synth. Biol. 11, 1292–1302 (2022).
Garabedian, M. V. et al. Designer membraneless organelles sequester native factors for control of cell behavior. Nat. Chem. Biol. 17, 998–1007 (2021). The first report to develop a synthetic membraneless organelle platform to control endogenous cellular activities by sequestration of native proteins.
Yoshikawa, M., Yoshii, T., Ikuta, M. & Tsukiji, S. Synthetic protein condensates that inducibly recruit and release protein activity in living cells. J. Am. Chem. Soc. 143, 6434–6446 (2021).
Montano Lopez, J., Duran, L. & Avalos, J. L. Physiological limitations and opportunities in microbial metabolic engineering. Nat. Rev. Microbiol. 20, 35–48 (2022).
Zhang, Y., Narlikar, G. J. & Kutateladze, T. G. Enzymatic reactions inside biological condensates. J. Mol. Biol. 433, 166624 (2021).
Huang, Z. et al. Rapid regulations of metabolic reactions in Escherichia coli via light-responsive enzyme redistribution. Biotechnol. J. 17, e2200129 (2022).
Peeples, W. & Rosen, M. K. Mechanistic dissection of increased enzymatic rate in a phase-separated compartment. Nat. Chem. Biol. 17, 693–702 (2021).
Song, D., Jo, Y., Choi, J. M. & Jung, Y. Client proximity enhancement inside cellular membrane-less compartments governed by client-compartment interactions. Nat. Commun. 11, 5642 (2020).
Na, D. et al. Metabolic engineering of Escherichia coli using synthetic small regulatory RNAs. Nat. Biotechnol. 31, 170–174 (2013).
Wang, Y. et al. Phase-separated multienzyme compartmentalization for terpene biosynthesis in a prokaryote. Angew. Chem. Int. Ed. 61, e202203909 (2022).
Reinkemeier, C. D., Girona, G. E. & Lemke, E. A. Designer membraneless organelles enable codon reassignment of selected mRNAs in eukaryotes. Science 363, eaaw2644 (2019). The first report to create an artificial orthogonally translating organelle enabling site-specific protein engineering with tailored noncanonical function.
Li, M. et al. Controlling synthetic membraneless organelles by a red-light-dependent singlet oxygen-generating protein. Nat. Commun. 13, 3197 (2022). The first report on red light-induced liquid-to-solid transition of synthetic protein condensates in cells.
Lasker, K. et al. The material properties of a bacterial-derived biomolecular condensate tune biological function in natural and synthetic systems. Nat. Commun. 13, 5643 (2022).
Alshareedah, I., Moosa, M. M., Pham, M., Potoyan, D. A. & Banerjee, P. R. Programmable viscoelasticity in protein-RNA condensates with disordered sticker-spacer polypeptides. Nat. Commun. 12, 6620 (2021).
Ghosh, A., Kota, D. & Zhou, H. X. Shear relaxation governs fusion dynamics of biomolecular condensates. Nat. Commun. 12, 5995 (2021).
Linsenmeier, M. et al. Dynamic arrest and aging of biomolecular condensates are modulated by low-complexity domains, RNA and biochemical activity. Nat. Commun. 13, 3030 (2022).
Liu, A. P. et al. The living interface between synthetic biology and biomaterial design. Nat. Mater. 21, 390–397 (2022).
Rodrigo-Navarro, A., Sankaran, S., Dalby, M. J., del Campo, A. & Salmeron-Sanchez, M. Engineered living biomaterials. Nat. Rev. Mater. 6, 1175–1190 (2021).
Tang, T.-C. et al. Materials design by synthetic biology. Nat. Rev. Mater. 6, 332–350 (2020).
Bracha, D., Walls, M. T. & Brangwynne, C. P. Probing and engineering liquid-phase organelles. Nat. Biotechnol. 37, 1435–1445 (2019). A systematic review on the state-of-the art technologies and approaches to probe and engineer condensates.
Fuxreiter, M. & Vendruscolo, M. Generic nature of the condensed states of proteins. Nat. Cell Biol. 23, 587–594 (2021).
Fisher, R. S. & Elbaum-Garfinkle, S. Tunable multiphase dynamics of arginine and lysine liquid condensates. Nat. Commun. 11, 4628 (2020).
Yoshikawa, M. & Tsukiji, S. Modularly built synthetic membraneless organelles enabling targeted protein sequestration and release. Biochemistry 60, 3273–3276 (2021).
Ladouceur, A. M. et al. Clusters of bacterial RNA polymerase are biomolecular condensates that assemble through liquid-liquid phase separation. Proc. Natl Acad. Sci. USA 117, 18540–18549 (2020).
Cohan, M. C. & Pappu, R. V. Making the case for disordered proteins and biomolecular condensates in bacteria. Trends Biochem. Sci. 45, 668–680 (2020).
Acknowledgements
This work is supported by the National Key Research and Development Program of China (grants 2020YFA0907702 and 2021YFA0909502 to X.-X.X.), the National Natural Science Foundation of China (grant 32071414 to X.-X.X. and grants 32270107 and 22075179 to Z.-G.Q.) and the Natural Science Foundation of Shanghai (grant 21ZR1432100 to Z.-G.Q.).
Author information
Authors and Affiliations
Contributions
X.-X.X. and Z.-G.Q. conceived of the manuscript. Z.-G.Q. and X.-X.X. wrote the paper. S.-C.H. reviewed the literature and edited the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Chemical Biology thanks G.-Q. Chen, F. Sun and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Qian, ZG., Huang, SC. & Xia, XX. Synthetic protein condensates for cellular and metabolic engineering. Nat Chem Biol 18, 1330–1340 (2022). https://doi.org/10.1038/s41589-022-01203-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-022-01203-3
This article is cited by
-
Assembling membraneless organelles from de novo designed proteins
Nature Chemistry (2024)
-
Programmable adhesion and morphing of protein hydrogels for underwater robots
Nature Communications (2024)
-
Local environment in biomolecular condensates modulates enzymatic activity across length scales
Nature Communications (2024)
-
Combinatorial optimization of gene expression through recombinase-mediated promoter and terminator shuffling in yeast
Nature Communications (2024)
-
Biomolecular condensates in kidney physiology and disease
Nature Reviews Nephrology (2023)