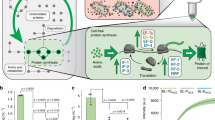
Abstract
Synthetic biology has shown remarkable potential to program living microorganisms for applications. However, a notable discrepancy exists between the current engineering practice—which focuses predominantly on planktonic cells—and the ubiquitous observation of microbes in nature that constantly alternate their lifestyles on environmental variations. Here we present the de novo construction of a synthetic genetic program that regulates bacterial life cycle and enables phase-specific gene expression. The program is orthogonal, harnessing an engineered protein from 45 candidates as the biofilm matrix building block. It is also highly controllable, allowing directed biofilm assembly and decomposition as well as responsive autonomous planktonic-biofilm phase transition. Coupling to synthesis modules, it is further programmable for various functional realizations that conjugate phase-specific biomolecular production with lifestyle alteration. This work establishes a versatile platform for microbial engineering across physiological regimes, thereby shedding light on a promising path for gene circuit applications in complex contexts.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Protein accession numbers (UniProt) are listed in Supplementary Table 1. Sequences for promoters and genes are provided in Supplementary Table 3. Source data are provided with this paper.
References
Cameron, D. E., Bashor, C. J. & Collins, J. J. A brief history of synthetic biology. Nat. Rev. Microbiol. 12, 381–390 (2014).
Endy, D. Foundations for engineering biology. Nature 438, 449–453 (2005).
Nandagopal, N. & Elowitz, M. B. Synthetic biology: integrated gene circuits. Science 333, 1244–1248 (2011).
Purnick, P. E. & Weiss, R. The second wave of synthetic biology: from modules to systems. Nat. Rev. Mol. Cell Biol. 10, 410–422 (2009).
Brophy, J. A. & Voigt, C. A. Principles of genetic circuit design. Nat. Methods 11, 508–520 (2014).
You, L., Cox, R. S., Weiss, R. & Arnold, F. H. Programmed population control by cell–cell communication and regulated killing. Nature 428, 868–871 (2004).
Tabor, J. J. et al. A synthetic genetic edge detection program. Cell 137, 1272–1281 (2009).
Danino, T., Mondragón-Palomino, O., Tsimring, L. & Hasty, J. A synchronized quorum of genetic clocks. Nature 463, 326–330 (2010).
Qi, L. S. et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183 (2013).
Mee, M. T., Collins, J. J., Church, G. M. & Wang, H. H. Syntrophic exchange in synthetic microbial communities. Proc. Natl Acad. Sci. USA 111, E2149–E2156 (2014).
Chen, Y., Kim, J. K., Hirning, A. J., Josić, K. & Bennett, M. R. Emergent genetic oscillations in a synthetic microbial consortium. Science 349, 986–989 (2015).
Kong, W., Meldgin, D. R., Collins, J. J. & Lu, T. Designing microbial consortia with defined social interactions. Nat. Chem. Biol. 14, 821–829 (2018).
Carothers, J. M., Goler, J. A. & Keasling, J. D. Chemical synthesis using synthetic biology. Curr. Opin. Biotechnol. 20, 498–503 (2009).
Smanski, M. J. et al. Synthetic biology to access and expand nature’s chemical diversity. Nat. Rev. Microbiol. 14, 135–149 (2016).
Gilbert, C. et al. Living materials with programmable functionalities grown from engineered microbial co-cultures. Nat. Mater. 20, 691–700 (2021).
Tang, T.-C. et al. Materials design by synthetic biology. Nat. Rev. Mater. 6, 332–350 (2021).
Rylott, E. L. & Bruce, N. C. How synthetic biology can help bioremediation. Curr. Opin. Chem. Biol. 58, 86–95 (2020).
de Lorenzo, V. Systems biology approaches to bioremediation. Curr. Opin. Biotechnol. 19, 579–589 (2008).
Ruder, W. C., Lu, T. & Collins, J. J. Synthetic biology moving into the clinic. Science 333, 1248–1252 (2011).
Weber, W. & Fussenegger, M. Emerging biomedical applications of synthetic biology. Nat. Rev. Genet. 13, 21–35 (2012).
Kitada, T., DiAndreth, B., Teague, B. & Weiss, R. Programming gene and engineered-cell therapies with synthetic biology. Science 359, eaad1067 (2018).
O’Toole, G., Kaplan, H. B. & Kolter, R. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54, 49–79 (2000).
Hall-Stoodley, L., Costerton, J. W. & Stoodley, P. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2, 95–108 (2004).
Flemming, H.-C. et al. Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol. 14, 563–575 (2016).
Prindle, A., Liu, J., Asally, M., Garcia-Ojalvo, J. & Süel, G. M. Ion channels enable electrical communication in bacterial communities. Nature 527, 59–63 (2015).
Flemming, H.-C. & Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 8, 623–633 (2010).
Alldredge, A. L. & Silver, M. W. Characteristics, dynamics and significance of marine snow. Prog. Oceanogr. 20, 41–82 (1988).
Azam, F. & Long, R. A. Sea snow microcosms. Nature 414, 495–498 (2001).
Zarrinpar, A., Chaix, A., Yooseph, S. & Panda, S. Diet and feeding pattern affect the diurnal dynamics of the gut microbiome. Cell Metab. 20, 1006–1017 (2014).
Marinangeli, C., Harding, S., Zafron, M. & Rideout, T. A systematic review of the effect of dietary pulses on microbial populations inhabiting the human gut. Benef. Microbes 11, 457–468 (2020).
Wood, T. K., Hong, S. H. & Ma, Q. Engineering biofilm formation and dispersal. Trends Biotechnol. 29, 87–94 (2011).
Lee, J., Jayaraman, A. & Wood, T. K. Indole is an inter-species biofilm signal mediated by SdiA. BMC Microbiol. 7, 42 (2007).
Hong, S. H. et al. Synthetic quorum-sensing circuit to control consortial biofilm formation and dispersal in a microfluidic device. Nat. Commun. 3, 613 (2012).
Qureshi, N., Annous, B. A., Ezeji, T. C., Karcher, P. & Maddox, I. S. Biofilm reactors for industrial bioconversion processes: employing potential of enhanced reaction rates. Microb. Cell Factories 4, 24 (2005).
Singh, R., Paul, D. & Jain, R. K. Biofilms: implications in bioremediation. Trends Microbiol. 14, 389–397 (2006).
Nicolella, C., Van Loosdrecht, M. & Heijnen, J. Wastewater treatment with particulate biofilm reactors. J. Biotechnol. 80, 1–33 (2000).
Jayaraman, A., Earthman, J. & Wood, T. Corrosion inhibition by aerobic biofilms on SAE 1018 steel. Appl. Microbiol. Biotechnol. 47, 62–68 (1997).
Tran, P. & Prindle, A. Synthetic biology in biofilms: tools, challenges, and opportunities. Biotechnol. Prog. 37, e3123 (2021).
Glass, D. S. & Riedel-Kruse, I. H. A synthetic bacterial cell-cell adhesion toolbox for programming multicellular morphologies and patterns. Cell 174, 649–658. e16 (2018).
Chen, B. et al. Programmable living assembly of materials by bacterial adhesion. Nat. Chem. Biol. 18, 289–294 (2022).
Trunk, T., Khalil, H. S. & Leo, J. C. Bacterial autoaggregation. AIMS Microbiol. 4, 140 (2018).
Kuipers, O. P., de Ruyter, P. G., Kleerebezem, M. & de Vos, W. M. Quorum sensing-controlled gene expression in lactic acid bacteria. J. Biotechnol. 64, 15–21 (1998).
Jinek, M. et al. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012).
Mu, D., Montalbán-López, M., Masuda, Y. & Kuipers, O. P. Zirex: a novel zinc-regulated expression system for Lactococcus lactis. Appl. Environ. Microbiol. 79, 4503–4508 (2013).
Llull, D. & Poquet, I. New expression system tightly controlled by zinc availability in Lactococcus lactis. Appl. Environ. Microbiol. 70, 5398–5406 (2004).
Brøndsted, L., Pedersen, M. & Hammer, K. An activator of transcription regulates phage TP901-1 late gene expression. Appl. Environ. Microbiol. 67, 5626–5633 (2001).
Paine, A., Eiz-Vesper, B., Blasczyk, R. & Immenschuh, S. Signaling to heme oxygenase-1 and its anti-inflammatory therapeutic potential. Biochem. Pharmacol. 80, 1895–1903 (2010).
Douglas, G. L. & Klaenhammer, T. R. Directed chromosomal integration and expression of the reporter gene gusA3 in Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 77, 7365–7371 (2011).
Shukla, T. P. & Wierzbicki, L. E. Beta‐galactosidase technology: a solution to the lactose problem. Crit. Rev. Food Sci. Nutr. 5, 325–356 (1975).
Marugg, J. D. et al. Cloning, expression, and nucleotide sequence of genes involved in production of pediocin PA-1, and bacteriocin from Pediococcus acidilactici PAC1.0. Appl. Environ. Microbiol. 58, 2360–2367 (1992).
Lu, W., Kong, W., Yang, P. & Kong, J. A one-step PCR-based method for specific identification of 10 common lactic acid bacteria and Bifidobacterium in fermented milk. Int. Dairy J. 41, 7–12 (2015).
Bairoch, A. et al. The universal protein resource (UniProt). Nucleic Acids Res. 33, D154–D159 (2005).
Cameron, D. E. & Collins, J. J. Tunable protein degradation in bacteria. Nat. Biotechnol. 32, 1276–1281 (2014).
Sumrin, A. et al. Purification and medium optimization of α-amylase from Bacillus subtilis 168. Afr. J. Biotechnol. 10, 2119–2129 (2011).
Zhang, K., Su, L. & Wu, J. Enhanced extracellular pullulanase production in Bacillus subtilis using protease-deficient strains and optimal feeding. Appl. Microbiol. Biotechnol. 102, 5089–5103 (2018).
O’Toole, G. A. Microtiter dish biofilm formation assay. J. Vis. Exp. 30, 2437 (2011).
Kong, W., Kapuganti, V. S. & Lu, T. A gene network engineering platform for lactic acid bacteria. Nucleic Acids Res. 44, e37 (2016).
Acknowledgements
This work was supported by the Office of Naval Research (grant no. N00014-16-1-2525).
Author information
Authors and Affiliations
Contributions
T.L. conceived the project. T.L. and P.S.S. designed the study. W.K. screened and characterized biofilm-forming proteins, constructed circuits for controllable biofilm formation and engineered P45 variants for biofilm dispersal. W.K. and Y.Q. developed platform for phase transition and function realization. W.K. and Y.Q. carried out experiments for auto-aggregation, biofilm quantification, product measurement and collected the data. W.K., Y.Q. and T.L. analyzed the data. T.L. and W.K. wrote the article with input from P.S.S. and Y.Q.
Corresponding author
Ethics declarations
Competing interests
T.L., Y.Q. and W.K. have filed a provisional patent application (application number: 63/404,971) on the platform described in this text. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Chemical Biology thanks Seok Hoon Hong and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Additional characterization of biofilm matrix proteins.
a, Plasmid for constitutive expression of the biofilm matrix proteins. b, Thickness of the biofilms formed on the surface of non-tissue culture treated 96-well plate by the library of 45 L. lactis strains that express predicted surface proteins (P1 to P45). c, SEM images of the biofilms formed by the strains encoding the proteins P6, P13, P25 and P40. Data are presented as mean ± s.d. from 3 independent experiments, and representative pictures from different samples are shown.
Extended Data Fig. 2 Dispersal of synthetic biofilms from plastic surfaces.
a, Protease-based dispersal of the biofilms made of P6, P25, P40 and P45. The biofilms on a polystyrene cell culture treated 96 well plate were directly quantified by crystal violet staining without any treatment, or treated by PBS or proteinase K (10 µg ml−1) for 2 hours at room temperature before being quantified. Data are presented as mean ± s.d. (n = 3 independent experiments). b, SEM images of intact, untreated biofilms and proteinase K-treated biofilms on polystyrene plastic sheets.
Extended Data Fig. 3 Additional characterizations of the P45 variants.
a, Quantification of biofilms formed on the polystyrene cell culture treated 96 well plate for the variants IS1-IS5. b, Images of test tubes containing the cultures of the variants at pH 7.4 and pH 5.0. c, Quantification of the aggregation ability of the variants at pH 7.4 and pH 5.0. For all panels, the strain P45 was used as a control. Data are presented as mean ± s.d. from n = 3 independent experiments. Representative pictures from different samples are shown.
Extended Data Fig. 4 Protease secretion and in vitro biofilm dispersal.
a, Protease secretion by L. lactis NZ9000 upon nisin induction. Lane 1, protein ladder. Lane 2, control without protease secretion. Lane 3 and 4, Protease A. Lane 5 and 6, Protease B. Lane 7 and 8, Protease C. Black arrow indicates the band of Usp45. Red arrow indicates Protease A. Green arrow indicates Protease B. Blue arrow indicates Protease C. The absence of the Usp45 band in Lane 5–8 suggests that Proteases B and C both exhibit proteolytic activity to digest Usp45. b, Inhibition of the IS5 biofilm by the supernatants of the protease-secreting strains. Overnight culture of IS5 was diluted with fresh medium to the OD600 of 0.04, then 120 µl of the diluted culture was added to a cell culture treated 96-well plate. 30 µl of L. lactis NZ9000 supernatants containing different proteases were added into the IS5 culture. Biofilm thickness was measured after growth for 24 hours. Data are presented as mean ± s.d. from 3 independent experiments.
Extended Data Fig. 5 Plasmid maps and control experiments for planktonic-biofilm transition.
a, Map of the plasmid IS5-Zn-gfp-prob. b, Map of the plasmid P45-Zn-gfp. c, Gene circuit of the plasmid P45-Zn-gfp. d-i, State transition experiments for the strain carrying the plasmid P45-Zn-gfp under different temporal patterns of zinc availability. Compared to the case of the strain carrying the plasmid IS5-Zn-gfp-prob (Fig. 4), the biofilm of the P45-Zn-gfp loaded strain cannot be decomposed once it forms. Experimental data are presented as mean ± s.d. from 3 independent experiments.
Extended Data Fig. 6 Increased antibiotic resistance coupled with biofilm formation.
a, Design of the gene circuit IS5-orf29-P7-Erm-Zn-gfp-prob. Building on the circuit IS5-Zn-gfp-prob, this system was established by introducing the transcriptional activator gene Orf29 at the downstream of IS5 and using the cognate promoter P7 to drive the expression of the erythromycin (Erm) resistance gene. b, Validations of the biofilm-coupled Erm resistance with colony forming unit counting. Cells containing the circuit IS5-orf29-P7-Erm-Zn-gfp-prob or the circuit IS5-Zn-gfp-prob were pre-cultured in the GM17/Cm/Zn media to be induced to the planktonic state or in the GM17/Cm/EDTA media to be induced to the biofilm state for 36 h with inoculations to fresh medium occurring every 12 h. Then, cell cultures with OD600 of 1.0 were serially diluted by 100–106 folds, and 5 µl of diluted cultures were added onto the agar plate supplemented with Cm to select all cells and the agar plate with Erm to select cells with the Erm resistance. c,d, State transitions of the strain carrying the circuit IS5-orf29-P7-Erm-Zn-gfp-prob under different temporal patterns of zinc availability. The Erm resistance was coupled with biofilm formation. e,f, State transition experiments for the control strain carrying IS5-Zn-gfp-prob under different temporal patterns of zinc availability. The Erm resistance remained low regardless of the life cycle. Data are presented as mean ± s.d. from 3 independent experiments.
Extended Data Fig. 7 Control experiments for coordinated lifestyle transition and amylase synthesis.
a-b, Quantification of the biofilm thickness and amylase activity of the amylase-encoding strain, which carries the plasmid IS5-Zn-amy-prob in the constant presence (a) and absence (b) of zinc. c-f, Quantification of the biofilm thickness and amylase activity of the strain carrying the plasmid P45-Zn-amy in four different zinc-changing environments. Experimental data are presented as mean ± s.d. from 3 independent experiments.
Extended Data Fig. 8 Control experiments for coordinated lifestyle transition and mHO-1 synthesis.
a-b, Quantification of the biofilm thickness and mHO-1 concentration of the mHO-1-encoding strain, which carries the plasmid IS5-Zn-mHO-1-prob in the constant presence (a) and absence (b) of zinc. c-f, Quantification of the biofilm thickness and mHO-1 concentration of the strain carrying the plasmid P45-Zn-mHO-1 in four different zinc-changing environments. Experimental data are presented as mean ± s.d. from 3 independent experiments.
Extended Data Fig. 9 Application of the lifestyle program for phase-specific, intracellular enzyme production.
a, Design of a gene circuit (IS5-Zn-gusA-prob) for GusA production by leveraging the modular structure in Fig. 5a. Here, the functional gene is gusA, which encodes beta-glucuronidase that converts p-nitrophenyl-β-D-glucopyranoside (PNPG) into the products, glucuronic acid and para-nitrophenol (PNP). PNP can be quantitatively measured by spectrometry at 420 nm. Compared to the functional molecules demonstrated in Fig. 5, one key difference here is that GusA remains intracellular and is not secreted to extracellular milieu. b-e, Quantification of the biofilm thickness and GusA activity of the strain carrying the plasmid IS5-Zn-gusA-prob in different zinc-changing environments. Notably, in response to zinc variations, cellular phase transitioned between the planktonic and biofilm states owing to the coordinated expression of IS5 and Protease B. However, there was no obvious reduction of GusA activity due to its high stability in the cell. f, Gene circuit for the plasmid P45-Zn-gusA. g-j, Quantification of the biofilm thickness and GusA activity of the strain carrying the plasmid P45-Zn-gusA in different zinc-changing environments. Neither biofilm decomposition nor GusA reduction was observed for this construct due to the lack of active degradation of P45 and GusA. Experimental data are presented as mean ± s.d. from 3 independent experiments.
Extended Data Fig. 10 Optimization of phase-specific control of intracellular GusA via engineered fast degradation.
a, Gene circuit for the optimized system, IS5-Zn-gusA-tag-prob-Pcst-lon, which contains an orthogonal protein degradation system (mf-Ion) and a degradation tag for GusA (gusA/tag). When zinc is present, IS5 expression is suppressed but Protease B is actively produced and secreted to disperse existing IS5 biofilm. Meanwhile, gusA is actively expressed with a fast degradation tag that can be recognized by the protease Mf-lon. In this case, the cell is in the planktonic state with a high level of tagged GusA. When zinc is absent, IS5 expression is turned on while the synthesis of Protease B is shut off, leading to biofilm formation. Meanwhile, the production of new GusA molecules is suppressed but the protease Mf-lon continues to actively digest existing tagged GusA, resulting in reduction of intracellular GusA concentration. The gene mf-lon is under the control of the low pH inducible promoter Pcst which is only active in the stationary phase, which reduces metabolic load and avoids excessive digestion of GusA when zinc is present. b-e, Quantification of the biofilm thickness and GusA activity of the strain carrying the plasmid IS5-Zn-gusA-tag-prob-Pcst-lon in different zinc-changing environments. With the optimized system, both cellular phase and GusA bioactivity showed clear transitions in response to environmental zinc availability. Experimental data are presented as mean ± s.d. from 3 independent experiments.
Supplementary information
Supplementary Information
Supplementary Figs. 1–3, Tables 1–3 and Refs. 1–6.
Supplementary Data 1
Source data for supplementary figures.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 4
Unprocessed gel.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 9
Statistical source data.
Source Data Extended Data Fig. 10
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kong, W., Qian, Y., Stewart, P.S. et al. De novo engineering of a bacterial lifestyle program. Nat Chem Biol 19, 488–497 (2023). https://doi.org/10.1038/s41589-022-01194-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-022-01194-1
This article is cited by
-
A change of phase
Nature Chemical Biology (2023)
-
Engineered autonomous dynamic regulation of metabolic flux
Nature Reviews Bioengineering (2023)
-
Diurnal switches in diazotrophic lifestyle increase nitrogen contribution to cereals
Nature Communications (2023)