Abstract
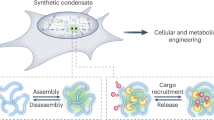
Subcellular compartmentalization of macromolecules increases flux and prevents inhibitory interactions to control biochemical reactions. Inspired by this functionality, we sought to build designer compartments that function as hubs to regulate the flow of information through cellular control systems. We report a synthetic membraneless organelle platform to control endogenous cellular activities through sequestration and insulation of native proteins. We engineer and express a disordered protein scaffold to assemble micron-size condensates and recruit endogenous clients via genomic tagging with high-affinity dimerization motifs. By relocalizing up to 90% of targeted enzymes to synthetic condensates, we efficiently control cellular behaviors, including proliferation, division and cytoskeletal organization. Further, we demonstrate multiple strategies for controlled cargo release from condensates to switch cells between functional states. These synthetic organelles offer a powerful and generalizable approach to modularly control cell decision-making in a variety of model systems with broad applications for cellular engineering.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All data supporting the findings of this study are included in the published article and its supplementary information files. Original data are available from the corresponding authors upon reasonable request. Source data are provided with this paper.
References
Good, M. C., Zalatan, J. G. & Lim, W. A. Scaffold proteins: hubs for controlling the flow of cellular information. Science 332, 680–686 (2011).
al-Mohanna, F. A., Caddy, K. W. & Bolsover, S. R. The nucleus is insulated from large cytosolic calcium ion changes. Nature 367, 745–750 (1994).
Burack, W. R. & Shaw, A. S. Signal transduction: hanging on a scaffold. Curr. Opin. Cell Biol. 12, 211–216 (2000).
Burack, W. R., Cheng, A. M. & Shaw, A. S. Scaffolds, adaptors and linkers of TCR signaling: theory and practice. Curr. Opin. Immunol. 14, 312–316 (2002).
Bhattacharyya, R. P., Reményi, A., Yeh, B. J. & Lim, W. A. Domains, motifs, and scaffolds: the role of modular interactions in the evolution and wiring of cell signaling circuits. Annu. Rev. Biochem. 75, 655–680 (2006).
Scott, J. D. & Pawson, T. Cell signaling in space and time: where proteins come together and when they’re apart. Science 326, 1220–1224 (2009).
Banani, S. F., Lee, H. O., Hyman, A. A. & Rosen, M. K. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298 (2017).
Choudhary, S., Quin, M. B., Sanders, M. A., Johnson, E. T. & Schmidt-Dannert, C. Engineered protein nano-compartments for targeted enzyme localization. PLoS ONE 7, e33342 (2012).
Dueber, J. E. et al. Synthetic protein scaffolds provide modular control over metabolic flux. Nat. Biotechnol. 27, 753–759 (2009).
Aumiller, W. M. Jr., Pir Cakmak, F., Davis, B. W. & Keating, C. D. RNA-based coacervates as a model for membraneless organelles: formation, properties, and interfacial liposome assembly. Langmuir 32, 10042–10053 (2016).
Zhao, E. M. et al. Light-based control of metabolic flux through assembly of synthetic organelles. Nat. Chem. Biol. 15, 589–597 (2019).
Reinkemeier, C. D., Girona, G. E. & Lemke, E. A. Designer membraneless organelles enable codon reassignment of selected mRNAs in eukaryotes. Science 363, eaaw2644 (2019).
Shin, Y. et al. Spatiotemporal control of intracellular phase transitions using light-activated optoDroplets. Cell 168, 159–171 (2017).
Bracha, D. et al. Mapping local and global liquid phase behavior in living cells using photo-oligomerizable seeds. Cell 175, 1467–1480 (2018).
Dine, E., Gil, A. A., Uribe, G., Brangwynne, C. P. & Toettcher, J. E. Protein phase separation provides long-term memory of transient spatial stimuli. Cell Syst. 6, 655–663 (2018).
Song, D., Jo, Y., Choi, J. M. & Jung, Y. Client proximity enhancement inside cellular membrane-less compartments governed by client–compartment interactions. Nat. Commun. 11, 5642 (2020).
Wei, S. P. et al. Formation and functionalization of membraneless compartments in Escherichia coli. Nat. Chem. Biol. 16, 1143–1148 (2020).
Schuster, B. S. et al. Controllable protein phase separation and modular recruitment to form responsive membraneless organelles. Nat. Commun. 9, 2985 (2018).
Elbaum-Garfinkle, S. et al. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc. Natl Acad. Sci. USA 112, 7189–7194 (2015).
Schuster, B. S. et al. Identifying sequence perturbations to an intrinsically disordered protein that determine its phase-separation behavior. Proc. Natl Acad. Sci. USA 117, 11421–11431 (2020).
Reed, E. H., Schuster, B. S., Good, M. C. & Hammer, D. A. SPLIT: stable protein coacervation using a light induced transition. ACS Synth. Biol. 9, 500–507 (2020).
Caldwell, R. M. et al. Optochemical control of protein localization and activity within cell-like compartments. Biochemistry 57, 2590–2596 (2018).
Cao, X., Jin, X. & Liu, B. The involvement of stress granules in aging and aging-associated diseases. Aging Cell 19, e13136 (2020).
Banani, S. F. et al. Compositional control of phase-separated cellular bodies. Cell 166, 651–663 (2016).
Piraner, D. I., Wu, Y. & Shapiro, M. G. Modular thermal control of protein dimerization. ACS Synth. Biol. 8, 2256–2262 (2019).
Iida, H. & Yahara, I. Specific early-G1 blocks accompanied with stringent response in Saccharomyces cerevisiae lead to growth arrest in resting state similar to the G0 of higher eucaryotes. J. Cell Biol. 98, 1185–1193 (1984).
Adams, A. E., Johnson, D. I., Longnecker, R. M., Sloat, B. F. & Pringle, J. R. CDC42 and CDC43, two additional genes involved in budding and the establishment of cell polarity in the yeast Saccharomyces cerevisiae. J. Cell Biol. 111, 131–142 (1990).
Woods, B., Kuo, C. C., Wu, C. F., Zyla, T. R. & Lew, D. J. Polarity establishment requires localized activation of Cdc42. J. Cell Biol. 211, 19–26 (2015).
Yoshida, S. et al. Polo-like kinase Cdc5 controls the local activation of Rho1 to promote cytokinesis. Science 313, 108–111 (2006).
Toenjes, K. A., Simpson, D. & Johnson, D. I. Separate membrane targeting and anchoring domains function in the localization of the S. cerevisiae Cdc24p guanine nucleotide exchange factor. Curr. Genet. 45, 257–264 (2004).
Botchkarev, V. V. Jr., Rossio, V. & Yoshida, S. The budding yeast Polo-like kinase Cdc5 is released from the nucleus during anaphase for timely mitotic exit. Cell Cycle 13, 3260–3270 (2014).
Sagot, I., Klee, S. K. & Pellman, D. Yeast formins regulate cell polarity by controlling the assembly of actin cables. Nat. Cell Biol. 4, 42–50 (2002).
Moseley, J. B. & Goode, B. L. The yeast actin cytoskeleton: from cellular function to biochemical mechanism. Microbiol. Mol. Biol. Rev. 70, 605–645 (2006).
Chesarone, M., Gould, C. J., Moseley, J. B. & Goode, B. L. Displacement of formins from growing barbed ends by bud14 is critical for actin cable architecture and function. Dev. Cell 16, 292–302 (2009).
Zhang, W. et al. Optogenetic control with a photocleavable protein, PhoCl. Nat. Methods 14, 391–394 (2017).
Lu, X. et al. Photocleavable proteins that undergo fast and efficient dissociation. Chemi. Sci. https://doi.org/10.1039/D1SC01059J (2021).
Tanimura, S. & Takeda, K. ERK signalling as a regulator of cell motility. J. Biochem. 162, 145–154 (2017).
Nakamura, H. et al. Intracellular production of hydrogels and synthetic RNA granules by multivalent molecular interactions. Nat. Mater. 17, 79–89 (2018).
Gordley, R. M., Bugaj, L. J. & Lim, W. A. Modular engineering of cellular signaling proteins and networks. Curr. Opin. Struct. Biol. 39, 106–114 (2016).
Gordley, R. M. et al. Engineering dynamical control of cell fate switching using synthetic phospho-regulons. Proc. Natl Acad. Sci. USA 113, 13528–13533 (2016).
Wu, C. Y., Roybal, K. T., Puchner, E. M., Onuffer, J. & Lim, W. A. Remote control of therapeutic T cells through a small molecule-gated chimeric receptor. Science 350, aab4077 (2015).
Toda, S., Blauch, L. R., Tang, S. K. Y., Morsut, L. & Lim, W. A. Programming self-organizing multicellular structures with synthetic cell–cell signaling. Science 361, 156–162 (2018).
Li, Y. et al. Modular construction of mammalian gene circuits using TALE transcriptional repressors. Nat. Chem. Biol. 11, 207–213 (2015).
Najem, J. S. et al. Assembly and characterization of biomolecular memristors consisting of ion channel-doped lipid membranes. J. Vis. Exp. https://doi.org/10.3791/58998 (2019).
Bashor, C. J., Helman, N. C., Yan, S. & Lim, W. A. Using engineered scaffold interactions to reshape MAP kinase pathway signaling dynamics. Science 319, 1539–1543 (2008).
Lau, Y. H., Giessen, T. W., Altenburg, W. J. & Silver, P. A. Prokaryotic nanocompartments form synthetic organelles in a eukaryote. Nat. Commun. 9, 1311 (2018).
Sigmund, F. et al. Bacterial encapsulins as orthogonal compartments for mammalian cell engineering. Nat. Commun. 9, 1990 (2018).
Giessen, T. W. et al. Large protein organelles form a new iron sequestration system with high storage capacity. eLife 8, e46070 (2019).
Li, P. et al. Phase transitions in the assembly of multivalent signalling proteins. Nature 483, 336–340 (2012).
Feric, M. et al. Coexisting liquid phases underlie nucleolar subcompartments. Cell 165, 1686–1697 (2016).
Dzuricky, M., Rogers, B. A., Shahid, A., Cremer, P. S. & Chilkoti, A. De novo engineering of intracellular condensates using artificial disordered proteins. Nat. Chem. 12, 814–825 (2020).
Chong, P. A., Vernon, R. M. & Forman-Kay, J. D. RGG/RG motif regions in RNA binding and phase separation. J. Mol. Biol. 430, 4650–4665 (2018).
Thompson, K. E., Bashor, C. J., Lim, W. A. & Keating, A. E. SYNZIP protein interaction toolbox: in vitro and in vivo specifications of heterospecific coiled-coil interaction domains. ACS Synth. Biol. 1, 118–129 (2012).
Haruki, H., Nishikawa, J. & Laemmli, U. K. The anchor-away technique: rapid, conditional establishment of yeast mutant phenotypes. Mol. Cell 31, 925–932 (2008).
Nishimura, K., Fukagawa, T., Takisawa, H., Kakimoto, T. & Kanemaki, M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat. Methods 6, 917–922 (2009).
Sambrook, J., Fritsch, E. F. & Maniatis, T. Molecular Cloning: A Laboratory Manual 2nd edn (Cold Spring Harbor Laboratory, 1989).
Guthrie, G. C. & Fink, G. Guide to yeast genetics and molecular biology. Methods Enzymol. 194, 1–863 (1991).
Longtine, M. S., Fares, H. & Pringle, J. R. Role of the yeast Gin4p protein kinase in septin assembly and the relationship between septin assembly and septin function. J. Cell Biol. 143, 719–736 (1998).
Anand, R., Memisoglu, G. & Haber, J. Cas9-mediated gene editing in Saccharomyces cerevisiae. Preprint at Protocol Exchange https://doi.org/10.1038/protex.2017.021a (2017).
Acknowledgements
We thank A. Kumar for sharing yeast strains, the E. Bi lab for yeast plasmids and technical support, B. Xia and H. Ahmed for cloning, H. Ramage for U2OS cell lines, the M. Shapiro lab for TlpA plasmids, the D. Hammer lab for critical reading of the manuscript and A. Stout and the Penn CDB Microscopy Core for imaging and support. This cellular-engineering study was supported by a National Institute of Biomedical Imaging and Bioengineering R01 grant EB028320 (M.C.G.). Biochemical characterization of disordered proteins was partly funded by a National Science Foundation (NSF) iSuperseed grant DMR1720530 (M.C.G.). Synthesis of optochemical dimerizers was supported by NSF grant CHE-1404836 (A.D.). Conceptual development of condensates as decision hubs and investigator salary support was supported, in part, by a Department of Energy BES Biomolecular Materials grant DE-SC0007063 (M.C.G.).
Author information
Authors and Affiliations
Contributions
M.V.G., J.B.D., W.W., R.M.C., W.B. and M.C.G. conceptualized the project and designed the experiments. J.B.D., M.T. and W.B. performed initial characterization of scaffold valency on condensate formation and client recruitment in yeast. M.V.G. performed cloning, strain generation and imaging for yeast experiments throughout the article. W.W. generated mammalian knock-in cell lines, imaged them and characterized client relocalization. B.S.S. contributed imaging data from yeast experiments included in Extended Data Fig. 1. A.D. contributed synthesized dimerizers. M.V.G., W.W. and M.C.G. analyzed data. M.V.G. and M.C.G. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Chemical Biology thanks Ulrich Krauss and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Properties of in vivo synthetic condensates.
a, Temperature dependence of condensate assembly as a function of scaffold RGG domain valency. Representative images of yeast cells expressing galactose induced GFP tagged scaffold with 1, 2, or 3 RGG domains at different temperatures. b, Heat map: quantitation of turbidity data of purified proteins from Schuster et al., 2018. c, Heat map: number of condensates per cell as a function of temperature and RGG domain valency. d, Fluorescence recovery after photobleaching (FRAP) of condensates formed by (RGG)3 scaffold; n = 10 condensates. Shaded area, 95% CI. e, Fluorescence loss in photobleaching (FLIP) of condensates formed by (RGG)3 scaffold. n = 13 cells. f, Steady state cytoplasmic scaffold concentration outside of condensates (Ccyto) as a function of cellular concentration (Ccell) for 30 cells per scaffold type. Dashed line, slope of 1. g, Average enrichment of scaffold protein in condensates for SZ1-(RGG)3 or TsCC(A)-(RGG)3. n = 164 and 97 condensates respectively. Error bars, s.d. h, Representative images of exogenously expressed mScarlet-SZ2 and mScarlet-TsCC(B) diffusely distributed in cytosol in the absence of condensates. i, Representative images of cells expressing FRB-(RGG)3 scaffold and mScarlet-FKBP as a client. The client is diffuse in the cytosol before the addition of Rap and concentrated in condensates after Rap addition. j, Quantitation of the fraction of client protein as in i localized to condensates over time after Rap addition. n = 15 cells. Shaded area 95% CI.
Extended Data Fig. 2 Condensate expression relocalizes tagged native clients and regulates cell growth.
a, Representative images of tagged, natively expressed Cdc24 show its cortical localization. b, Representative images of tagged natively expressed Cdc5 show its punctate localization to spindle pole body. c, Images of the same Cdc24-mScarlet-TsCC(B) cell before and after induced expression of TsCC(A)-(RGG)3 scaffold for 6 hr, show loss of cortical Cdc24 signal and partitioning to synthetic condensate. d, Client recruitment to condensates specifically depends on CC tag interaction; Cdc24 does not interact with (RGG)3 condensates that lack the interaction tag. e, Left, scheme: cortical Cdc24 is relocalized from cortex to synthetic condensates after induction of scaffold. Right, Average cortical Cdc24- mScarlet-TsCC(B) signal before and after TsCC(A)-(RGG)3 scaffold expression (6 hr). n = 20 cells before and after hours of galactose induction. Significance calculated by unpaired, two-tailed, t-test (****, p < 0.0001). f, Kinetics of loss of cortical Cdc24-mCherry-TsCC(B) signal (red) concomitant with cellular accumulation of expressed TsCC(A)-(RGG)3 scaffold (green) upon induction with galactose for 20 cells over 6 hours. Shaded area, s.d. g, Cell proliferation: measurements of cell density (OD600) over time for indicated Cdc24 strains in liquid media containing galactose. h, Growth assay for Cdc24 strains: five-fold serial dilution of indicated strains grown on solid- media containing glucose or galactose. i, Average cell area of mother cells only increases upon TsCC(A)-(RGG)3 expression in Cdc24-mScarlet-TsCC(B) cells, consistent with cell cycle arrest in G1. j, Growth assay for Cdc5 strains: five-fold serial dilution of indicated strains grown on solid-media containing glucose or galactose. In all cases, growth defect depends on presence of tagged client and expression of scaffold to form condensates. Phenotype is not observed with only native client tagging or only scaffold expression.
Extended Data Fig. 3 Reversible control of cell proliferation-arrest state.
a, Representative images of Cdc24-mScarlet-TsCC(B) cells expressing TsCC(A)-(RGG)3 scaffold at the indicated temperatures for 14 hours. Thermally responsive coiled-coil pair dissociate upon heating to 37 or 42 °C, releasing client to promote cell polarity and reversing the cell cycle arrest associated with Cdc24 sequestration to condensates. b, Representative images of Cdc24-mScarlet-TsCC(B) in the presence of TsCC(A)-PhoCl2f-(RGG)3 before and after illumination with 405 nm light. c, Schematic of client release strategy: Cdc24 is tagged with PhoCl-TsCC(B). 405 nm light results in PhoCl cleavage and client release. d, Percentage of cells expressing Cdc24-mScarlet-TsCC(B) arrested (unbudded cells) over time after scaffold induction +/− illumination. n = 4048 cells in total pooled from three trials. e, Prediction: cycling of cell state between budded-arrested-budded-arrested. f, Representative images of cells at the indicated time points. Wildtype levels of budding at time 0 h. Cells incubated in galactose at 25 °C from 0-6 h timepoints to induce condensate formation, blocking budding, then heated from 6 h to 8 h timepoints, promoting polarization and budding and cooled back to 25 °C and arrested by 12 h.
Extended Data Fig. 4 Sequestration to synthetic condensates in mammalian cells.
a, Schematic of CRISPR tagging approach to endogenous loci in mammalian cells and expression of the scaffold by a CMV promoter. b, PCR validation of CRISPR tagging in mammalian cell lines. Only tagged strains show a PCR product of the expected size as indicated. c, Representative images of tagged ERK1 robustly partitions to synthetic condensates formed by expression of TsCC(A)-(RGG)3 scaffold. d, Representative images of tagged Par6 localized to the cell cortex in the absence of scaffold expression (left) and to condensate structures when scaffold is expressed (right). e, Quantitation of cortical Par6-mCherry-TsCC(B) in the absence and presence of condensates with cognate coiled coil. n = 10 cells for each condition. Error bars, s.d. Significance calculated by unpaired, two-tailed t-test. (**, p < 0.01).
Supplementary information
Supplementary Information
Supplementary Tables 1 and 2.
Supplementary Video 1
Video 1. Rapamaycin-induced client recruitment to preformed condensates in cells; green, FRB–(RGG)3–GFP-tagged scaffold; red, exogenous client, mScarlet–TsCC(B).
Supplementary Video 2
Video 2. Inducible expression of scaffold disrupts native cortical localization of Cdc24, sequestering it to newly formed synthetic condensates; green, TsCC(A)–(RGG)3–GFP-tagged scaffold; red, native client, Cdc24–mScarlet–TsCC(B).
Supplementary Video 3
Video 3. Induced condensate sequestration of endogenous client, Cdc24, following rapamycin addition; green, FRB–(RGG)3–GFP-tagged scaffold; red, native client, Cdc24–mScarlet–FKBP.
Source data
Source Data Fig. 3
Unprocessed images of spot assay plates.
Source Data Extended Data Fig. 2
Unprocessed images of spot assay plates.
Source Data Extended Data Fig. 4
Unprocessed gel image.
Rights and permissions
About this article
Cite this article
Garabedian, M.V., Wang, W., Dabdoub, J.B. et al. Designer membraneless organelles sequester native factors for control of cell behavior. Nat Chem Biol 17, 998–1007 (2021). https://doi.org/10.1038/s41589-021-00840-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-021-00840-4
This article is cited by
-
Local environment in biomolecular condensates modulates enzymatic activity across length scales
Nature Communications (2024)
-
Determinants that enable disordered protein assembly into discrete condensed phases
Nature Chemistry (2024)
-
Biomolecular condensates: insights into early and late steps of the HIV-1 replication cycle
Retrovirology (2023)
-
Microbial cell factories based on filamentous bacteria, yeasts, and fungi
Microbial Cell Factories (2023)
-
Programmable synthetic biomolecular condensates for cellular control
Nature Chemical Biology (2023)