Abstract
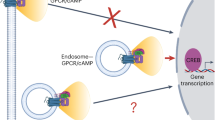
G-protein-coupled receptor-regulated cAMP production from endosomes can specify signaling to the nucleus by moving the source of cAMP without changing its overall amount. How this is possible remains unknown because cAMP gradients dissipate over the nanoscale, whereas endosomes typically localize micrometers from the nucleus. We show that the key location-dependent step for endosome-encoded transcriptional control is nuclear entry of cAMP-dependent protein kinase (PKA) catalytic subunits. These are sourced from punctate accumulations of PKA holoenzyme that are densely distributed in the cytoplasm and titrated by global cAMP into a discrete metastable state, in which catalytic subunits are bound but dynamically exchange. Mobile endosomes containing activated receptors collide with the metastable PKA puncta and pause in close contact. We propose that these properties enable cytoplasmic PKA to act collectively like a semiconductor, converting nanoscale cAMP gradients generated from endosomes into microscale elevations of free catalytic subunits to direct downstream signaling.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Kwok-Keung Fung, B. & Stryer, L. Photolyzed rhodopsin catalyzes the exchange of GTP for bound GDP in retinal rod outer segments. Proc. Natl Acad. Sci. USA 77, 2500–2504 (1980).
Tsvetanova, N. G., Irannejad, R. & von Zastrow, M. G protein-coupled receptor (GPCR) signaling via heterotrimeric G proteins from endosomes. J. Biol. Chem. 290, 6689–6696 (2015).
Pavlos, N. J. & Friedman, P. A. GPCR signaling and trafficking: the long and short of it. Trends Endocrinol. Metab. 28, 213–226 (2017).
Calebiro, D. & Godbole, A. Internalization of G-protein-coupled receptors: implication in receptor function, physiology and diseases. Best. Pract. Res. Clin. Endocrinol. Metab. 32, 83–91 (2018).
Vilardaga, J. P., Jean-Alphonse, F. G. & Gardella, T. J. Endosomal generation of cAMP in GPCR signaling. Nat. Chem. Biol. 10, 700–706 (2014).
Lohse, M. J. & Hofmann, K. P. Spatial and temporal aspects of signaling by G-protein-coupled receptors. Mol. Pharmacol. 88, 572–578 (2015).
Lobingier, B. T. & von Zastrow, M. When trafficking and signaling mix: how subcellular location shapes G protein-coupled receptor activation of heterotrimeric G proteins. Traffic 20, 130–136 (2019).
Calebiro, D. et al. Persistent cAMP-signals triggered by internalized G-protein-coupled receptors. PLoS Biol. 7, e1000172 (2009).
Ferrandon, S. et al. Sustained cyclic AMP production by parathyroid hormone receptor endocytosis. Nat. Chem. Biol. 5, 734–742 (2009).
Wehbi, V. L. et al. Noncanonical GPCR signaling arising from a PTH receptor-arrestin- Gβγ complex. Proc. Natl Acad. Sci. USA 110, 1530–1535 (2013).
Thomsen, A. R. B. et al. GPCR-G protein-beta-arrestin super-complex mediates sustained G protein signaling. Cell 166, 907–919 (2016).
Keenan, D. M. & Veldhuis, J. D. Pulsatility of hypothalamo-pituitary hormones: a challenge in quantification. Physiology 31, 34–50 (2016).
Lyga, S. et al. Persistent cAMP signaling by internalized LH receptors in ovarian follicles. Endocrinology 157, 1613–1621 (2016).
Irannejad, R. et al. Conformational biosensors reveal GPCR signalling from endosomes. Nature 495, 534–538 (2013).
Cao, T. T., Deacon, H. W., Reczek, D., Bretscher, A. & von Zastrow, M. A kinase-regulated PDZ-domain interaction controls endocytic sorting of the β2-adrenergic receptor. Nature 401, 286–290 (1999).
Uchida, Y., Rutaganira, F. U., Jullié, D., Shokat, K. M. & Von Zastrow, M. Endosomal phosphatidylinositol 3-kinase is essential for canonical GPCR signaling. Mol. Pharmacol. 91, 65–73 (2016).
Tsvetanova, N. G. & von Zastrow, M. Spatial encoding of cyclic AMP signaling specificity by GPCR endocytosis. Nat. Chem. Biol. 10, 1061–1065 (2014).
O’Banion, C. P., Vickerman, B. M., Haar, L. & Lawrence, D. S. Compartmentalized cAMP generation by engineered photoactivated adenylyl cyclases. Cell Chem. Biol. 26, 1393–1406.e7 (2019).
Lazar, A. M. et al. G protein-regulated endocytic trafficking of adenylyl cyclase type 9. eLife 9, e58039 (2020).
Binkowski, B. F. et al. A luminescent biosensor with increased dynamic range for intracellular cAMP. ACS Chem. Biol. 6, 1193–1197 (2011).
Gonzalez, G. A. & Montminy, M. R. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell 59, 675–680 (1989).
Nigg, E. A. A., Hilz, H., Eppenberger, H. M. M. & Dutly, F. Rapid and reversible translocation of the catalytic subunit of cAMP-dependent protein kinase type II from the Golgi complex to the nucleus. EMBO J. 4, 2801–2806 (1985).
Clister, T. et al. AKAP95 organizes a nuclear microdomain to control local cAMP for regulating nuclear PKA. Cell Chem. Biol. 26, 885–891.e4 (2019).
Kamiyama, D. et al. Versatile protein tagging in cells with split fluorescent protein. Nat. Commun. 7, 11046 (2016).
Leonetti, M. D. et al. A scalable strategy for high-throughput GFP tagging of endogenous human proteins. Proc. Natl Acad. Sci. USA 113, E3501–E3508 (2016).
Uhler, M. D., Chrivia, J. C. & McKnight, G. S. Evidence for a second isoform of the catalytic subunit of cAMP-dependent protein kinase. J. Biol. Chem. 261, 15360–15363 (1986).
Feng, S. et al. Improved split fluorescent proteins for endogenous protein labeling. Nat. Commun. 8, 370 (2017).
Mavillard, F., Hidalgo, J., Megias, D., Levitsky, K. L. & Velasco, A. PKA-mediated Golgi remodeling during cAMP signal transmission. Traffic 11, 90–109 (2010).
Sprague, B. L. & McNally, J. G. FRAP analysis of binding: proper and fitting. Trends Cell Biol. 15, 84–91 (2005).
Walker-Gray, R., Stengel, F. & Gold, M. G. Mechanisms for restraining cAMP-dependent protein kinase revealed by subunit quantitation and cross-linking approaches. Proc. Natl Acad. Sci. USA 114, 10414–10419 (2017).
Cheng, X., Phelps, C. & Taylor, S. S. Differential binding of cAMP-dependent protein kinase regulatory subunit isoforms Iα and IIβ to the catalytic subunit. J. Biol. Chem. 276, 4102–4108 (2001).
Zawadzki, K. M. & Taylor, S. S. cAMP-dependent protein kinase regulatory subunit type IIbeta: active site mutations define an isoform-specific network for allosteric signaling by cAMP. J. Biol. Chem. 279, 7029–7036 (2004).
Schlosshauer, M. & Baker, D. Realistic protein–protein association rates from a simple diffusional model neglecting long-range interactions, free energy barriers, and landscape ruggedness. Protein Sci. 13, 1660–1669 (2004).
Irannejad, R. et al. Functional selectivity of GPCR-directed drug action through location bias. Nat. Chem. Biol. 13, 799–806 (2017).
Gillooly, D. J. et al. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 19, 4577–4588 (2000).
Berg, H. C. & Purcell, E. M. Physics of chemoreception. Biophys. J. 20, 193–219 (1977).
Ashcroft, N. W. & Mermin, N. D. Solid State Physics (Harcourt, Inc., 1976).
Sze, S. M. Physics of Semiconductor Devices (John Wiley & Sons, Inc., 1981).
Zhang, J. Z. et al. Phase separation of a PKA regulatory subunit controls cAMP compartmentation and oncogenic signaling. Cell 182, 1531–1544.e15 (2020).
Bock, A. et al. Optical mapping of cAMP signaling at the nanometer scale. Cell 182, 1–12 (2020).
Beavo, J. A., Bechtel, P. J. & Krebs, E. G. Activation of protein kinase by physiological concentrations of cyclic AMP. Proc. Natl Acad. Sci. USA 71, 3580–3583 (1974).
Koschinski, A. & Zaccolo, M. Activation of PKA in cell requires higher concentration of cAMP than in vitro: implications for compartmentalization of cAMP signalling. Sci. Rep. 7, 1–12 (2017).
Zhao, Y. et al. An expanded palette of genetically encoded Ca2+ indicators. Science 333, 1888–1891 (2011).
Patriarchi, T. et al. Imaging neuromodulators with high spatiotemporal resolution using genetically encoded indicators. Nat. Protoc. 14, 3471–3505 (2019).
Niwa, H., Yamamura, K. & Miyazaki, J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108, 193–199 (1991).
Lin, S., Staahl, B. T., Alla, R. K. & Doudna, J. A. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife 3, e04766 (2014).
Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012).
Edelstein, A., Amodaj, N., Hoover, K., Vale, R. & Stuurman, N. Computer control of microscopes using µManager. Curr. Protoc. Mol. Biol. 92, 14.20.1–14.20.17 (2010).
Edelstein, A. D. et al. Advanced methods of microscope control using μManager software. J. Biol. Methods 1, e10 (2014).
Abràmoff, M. D., Magalhães, P. J. & Ram, S. J. Image processing with ImageJ. Biophotonics Int. 11, 36–42 (2004).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Acknowledgements
We thank A. Marley for developing the cAMP fluorescence biosensor and S. Kotowski for generating the DsRed-tagged dynamin1 constructs. We thank B. Lobingier for help with the development of the cAMP luminescence assay. We also thank N. Tsvetanova, R. Irannejad, D. Jullié and members of the von Zastrow laboratory for valuable advice and discussion, as well as K. Thorn, D. Larsen and K. Herrington (Nikon Imaging Center, UCSF, National Institute of Health 1S10OD017993-01A1) for technical support and expertise. We also thank M. Chevalier, R. Dror, N. L’Etoile, R. D. Mullins, S. Taylor, O. Weiner and J. Zhang for valuable discussion. These studies were supported by the National Institute on Drug Abuse (grant nos. DA012864 and DA010711 to M.v.Z.), the National Institute of Mental Health (grant no. MH120212 to M.v.Z.), the National Heart, Lung and Blood Institute (grant no. HL129689 to G.E.P.), the National Institute of Biomedical Imaging and Bioengineering (grant no. EB022798 to B.H.) and the American Heart Association (grant nos. 15PRE21770003 to V.P. and 16PRE26420057 to G.E.P.).
Author information
Authors and Affiliations
Contributions
G.E.P. and M.v.Z. designed the study and interpreted results. G.E.P. performed the experiments and analyzed data. V.P. and B.H. generated the endogenously labeled mNG2-PKAcat HEK293T cell line. G.E.P. and M.v.Z. wrote the manuscript, with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Characterization of endocytic blockade on β2AR and the effects of endocytic blockade on the cAMP signaling pathway.
a, cAMP luminescence time course from Fig. 1a (n = 3 biological replicates; control vs Dyngo4a mock p = 0.0469; control mock vs Iso, Dyngo4a mock vs Iso and control vs Dyngo4a Iso, p < 0.0001). b and c, Quantitative RT-PCR was performed on cells transfected with (b) mCherry-Dyn1 (control) or mCherry-Dyn1-K44E, or (c) Control siRNA or CHC17 siRNA, then untreated or treated with 100 nM Iso. PCK1 and GAPDH transcript levels determined by qRT-PCR. Dyn1 vs Dyn1K44E (n = 6 biological replicates, Interaction = 0.6313, Time p < 0.0001, Transfection p = 0.0476). Control siRNA vs CHC17 siRNA (n = 6 biological replicates, Interaction p = 0.0260, Time p < 0.0001, Transfection p < 0.0001). d, Iso stimulates cAMP luminescence over time. Cells were untreated (mock) or treated with 100 nM, 10 nM, or 1 nM Iso at 0 minutes. (n = 3 biological replicates; Interaction, Time and Treatment p < 0.0001; mock vs 1 nM Iso, mock vs 10 nM Iso and mock vs 100 nM Iso p < 0.0001). e, Maximum cAMP luminescence from d (n = 3 biological replicates; mock vs 1 nM Iso p = 0.7889; 10 nM vs 100 nM Iso, p = 0.0004; mock vs 100 nM Iso and 1 nM vs 100 nM Iso p < 0.0001). f, Induction of PCK1 increases with concentration of Iso. qRT-PCR was performed on cells untreated (mock) and treated with 100 nM, 10 nM, and 1 nM Iso (n = 3 biological replicates; mock vs 1 nM Iso, p = 0.2323; mock vs 100 nM Iso, p < 0.0001; 1 nM vs 100 nM Iso, p = 0.0006; 10 nM vs 100 nM Iso, p = 0.1270). g, Comparison of cAMP production using different doses of Iso and endocytic blockade with mutant dynamin. GG4B and pcDNA3 (control) or mCherry-Dyn1-K44E expressing cells were untreated (mock) or treated with 100 or 10 nM Iso. Area under the curve (right) shown for the quantification of the time course (left) (n = 4, cells ≥ 9 per biological replicate; control 10 nM Iso vs. Dyn1-K44E 100 nM Iso p = 0.3350; all other comparisons p < 0.0001). h, Kinetics of cAMP response. Each cAMP response from Fig. 1d was normalized to the peak. (control, n = 4, Dyn1-K44E n = 3, Interaction p = 0.9776, Time p < 0.0001, Transfection p = 0.0634). All data are mean ± sem (shaded areas). Significance determined by ordinary one-way ANOVA (e-g), or two-way ANOVA (a-d, h) with Tukey’s (a, d) or Sidak’s (c, e-g) multiple comparisons tests.
Extended Data Fig. 2 Endocytosis blockade reduces β2AR-stimulated cAMP downstream signaling.
a-c, Nuclear cAMP signaling cascade responses are shown in the same time scale (up to 60 minutes) in cells transfected with control siRNA or CHC17 siRNA. a, Western blot analysis of PKAcat from nuclear samples treated with 100 nM Iso (adapted from Fig. 2c,d). b, Western blot analysis of CREB phosphorylation from whole cell lysates after 100 nM Iso treatment (adapted from Fig. 2b). c, qRT-PCR quantitation of cells treated with 100 nM Iso (adapted from Extended Data Fig. 1c). d, Western blot quantification of PKAcat from whole cell lysate and nuclear samples (n = 3 biological replicates, p < 0.0001, two-tailed unpaired t-test). All data are mean ± sem.
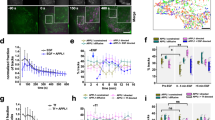
Extended Data Fig. 3 Detection of PKAcat by live microscopy gene-edited mNG2-PKAcat HEK293T cells.
a, Identifying cells expressing NLS-mNG21-10 IRES TagBFP for analysis. Images correspond with cells in b. b, Representative spinning disk confocal images of live cells expressing mNG211-PKAcat, mNG21-10 and nuclear localized NLS-mNG21-10. Cells were untreated (mock) or treated with 100 nM Iso at 0 minutes. Inset shows nuclear ROI shown as dotted line square. c and d, Quantification of PKAcat nuclear accumulation. c, Cells were untreated (mock) or stimulated with 100 nM Iso at 0 minutes in c (n = 3, cells ≥ 21 per biological replicate; Interaction and Time p = 0.0346, Treatment p < 0.0001; p < 0.05 mock vs Iso time = 5-35 and 45 minutes with Sidak’s multiple comparisons test; two-way ANOVA). d, Cells expressing pcDNA3 (control) or mCherry-Dyn1-K44E were untreated (mock) or treated with 100 nM Iso at 0 minutes. (n = 4, cells ≥ 7 per biological replicate; Interaction p = 0.7938, Time and Transfection p < 0.0001, two-way ANOVA). e and f, Identifying cells for analysis. mNG2-PKAcat cells expressing NLS-mNG21-10 IRES TagBFP were co-transfected with NLS-mNG21-10 IRES TagBFP and control or Dyn1-K44E DNA (e) or CHC17 siRNA (f). e, Images correspond to cells in Fig. 3c. g, Quantification of PKAcat nuclear accumulation in mNG2-PKAcat cells transfected with ASN siRNA (control) or AF555-CHC17 siRNA (n = 5, cells ≥ 9 per biological replicate; Interaction, Time and Transfection p < 0.0001; Control siRNA vs CHC17 siRNA mock p = 0.8237, all other comparisons p < 0.0001; Sidak’s multiple comparisons test, two-way ANOVA). h, Quantification of cells transiently expressing ASN siRNA (control) or AF555-CHC17 siRNA from g, data normalized to the corresponding untreated 45 minute time point (n = 5 biological replicates; p = 0.0017 Control siRNA untreated vs Iso, p = 0.5632 CHC17 siRNA untreated vs Iso, p = 0.03 Control siRNA Iso vs CHC17 siRNA Iso; Sidak’s multiple comparisons test, ordinary one-way ANOVA). i, Validation of CHC17 knockdown in imaging experiments quantified in g and h. qRT-PCR for CHC17 and GAPDH transcript levels (n = 5 biological replicates, p < 0.0001, two-tailed unpaired t-test). All data are mean ± sem (shaded areas). Scale bars = 5 μm.
Extended Data Fig. 4 Identification of cells expressing mCherry-Dyn1-K44Ein HEK293T mNG2-PKAcat cells.
Cells expressing mCherry-Dyn1-K44E were identified to determine which cells would be used for analysis. Images correspond with image of cells from Fig. 4b. Scale bar = 5 μm.
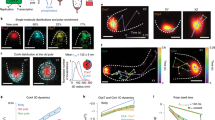
Extended Data Fig. 5 Fluorescence recovery after photobleaching analysis.
a, Quantification from Fig. 5c without normalizing to whole cell fluorescence. b, Photobleaching recovery curves with non-linear fit. Recovery curves from Fig. 5c are replotted without pre-photobleaching. Each recovery curve was fit using non-linear regression and an exponential one-phase association model. The Iso condition (blue) has a half-time of 50.40 s and fractional recovery of 86.04%. The untreated condition (gray) has a half-time of 58.86 s and a fractional recovery of 61.08% (n = 3, cells ≥ 3 per biological replicate; Interaction, Time and Transfection p < 0.0001; p < 0.05 for untreated vs Iso for time = 30-300 s with Sidak’s multiple comparisons test, two-way ANOVA). c, Photobleaching in different regions of the cell. Cells untreated or treated with 100 nM Iso for 30 minutes are photobleached in a perinuclear or cytoplasmic region (n = 3, cells ≥ 3 per biological replicate; Interaction, Time and Transfection p < 0.0001; all comparisons p < 0.0001 with Sidak’s multiple comparisons test, two-way ANOVA). d and e, Half times and percent maximal recovery of all photobleaching conditions determined by a non-linear regression and an exponential one-phase association model. d, Half times (n ≥ 3, cells ≥ 3 per biological replicate; p = 0.9645, ordinary one-way ANOVA). e, Percent maximal recovery (n ≥ 3, cells ≥ 3 per biological replicate; mock vs Iso and Iso vs Iso Alp p < 0.0001; Dyn1-K44E mock vs Iso and Iso vs Iso Alp p = 0.0005; DMSO vs Fsk p = 0.0057; Perinuclear ROI vs Cytoplasmic ROI p = 0.3736; Iso Alp vs Iso CGP p = 0.9847; Cyto mock vs Iso p = 0.9988; control vs Dyn1-K44E Iso and Iso vs Fsk p > 0.9999; Sidak’s multiple compariosons test, ordinary one-way ANOVA). f and g, Validation of PKAcat return to the perinuclear region after 100 nM forskolin (Fsk). f, Quantification of perinuclear PKAcat 30 minutes after DMSO, 100 nM, 10 μM and 10 μM Fsk treatment (n = 3, cells ≥ 20 per biological replicate; DMSO vs 10 nM Fsk p=0.9995; DMSO vs 100 nM Fsk p=0.9997; DMSO vs 1 μM Fsk p=0.7704; DMSO vs 10 μM Fsk p=0.0017; Sidak’s multiple comparisons test, ordinary one-way ANOVA). g, Representative live cell spinning disk confocal images of mNG2-PKAcat before and after 30 minute treatment with DMSO and 100 nM Fsk. Scale bar = 5 μm. Data are mean ± sem.
Extended Data Fig. 6 Endocytosis moves activated β2ARs into close proximity with dynamic PKAcat.
a, Cells were transfected with FLAG-β2AR and Nb80-mApple. Surface FLAG-β2ARs were labeled with M1-FLAG-647 for 15 minutes prior to imaging. Representative spinning disk confocal images of live cells were taken before and 30 minutes after 100 nM Iso addition. b, Spinning disk confocal fixed images of mNG2-PKAcat cells stained for EEA1. Individual channels from merged image in Fig. 6d. c, Spinning disk confocal live images of mNG2-PKAcat cells transiently expressing DsRed-EEA1. Cells were pretreated with 100 nM Iso for 25 minutes and imaged for 5 minutes at 5 second intervals. Scale bar = 5 μm.
Supplementary information
Supplementary Information
Supplementary Figs. 1–5 and Tables 1–7.
Supplementary Video 1
Movie of a live confocal image series of endogenously labeled mNG2-PKAcat (green) and endosomes marked by DsRed-EEA1 (2× FYVE) (magenta), providing a representative example of sequential dispersal and punctate relocalization of PKAcat during the biphasic β2AR response. A 25 min series is shown, with image capture every 20 s. Elapsed time is indicated at the upper left (mm:ss). 100 nM Iso was bath applied at 2:40. Scale bar, 5 µm.
Supplementary Video 2
Movie of fluorescence recovery after photobleaching of an untreated endogenously labeled mNG2-PKAcat cell. Cells were untreated for 30 min before imaging and photobleached at 30 s. Recovery was observed for 300 s after photobleaching. Corresponds to images in Fig. 5b, top row. Scale bar, 5 µm.
Supplementary Video 3
Movie of fluorescence recovery after photobleaching of an Iso-treated endogenously labeled mNG2-PKAcat cell. Cells were treated for 30 min with 100 nM Iso before imaging, and photobleached at 30 s. Recovery was observed for 300 s after photobleaching. Corresponds to images in Fig. 5b, bottom row. Scale bar, 5 µm.
Supplementary Video 4
Movie of a live confocal image series (corresponding with Extended Data Fig. 6c) of endogenously labeled mNG2-PKAcat (green and endosomes marked by DsRed-EEA1 (2× FYVE)) (magenta). A 50 s series is shown, with image capture every 5 s. The green channel displays endogenously labeled mNG2-PKAcat. The magenta channel displays endosomes marked by DsRed-EEA1 (2× FYVE). A calculated image corresponding to the product of the green and magenta channels is overlaid and displayed using the built-in ‘fire’ lookup table of ImageJ. Scale bar, 5 µm.
Supplementary Table 8
Statistical source data file for Supplementary Fig. 2.
Source data
Source Data Fig. 1
statistical source data.
Source Data Fig. 2
Unprocessed western blots.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Rights and permissions
About this article
Cite this article
Peng, G.E., Pessino, V., Huang, B. et al. Spatial decoding of endosomal cAMP signals by a metastable cytoplasmic PKA network. Nat Chem Biol 17, 558–566 (2021). https://doi.org/10.1038/s41589-021-00747-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-021-00747-0
This article is cited by
-
Endosome positioning coordinates spatially selective GPCR signaling
Nature Chemical Biology (2024)
-
Non-canonical β-adrenergic activation of ERK at endosomes
Nature (2022)
-
Location bias contributes to functionally selective responses of biased CXCR3 agonists
Nature Communications (2022)
-
Schwann cell endosome CGRP signals elicit periorbital mechanical allodynia in mice
Nature Communications (2022)