Abstract
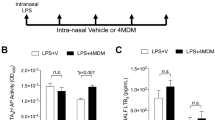
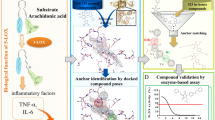
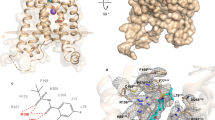
Leukotrienes (LT) are lipid mediators of the inflammatory response that are linked to asthma and atherosclerosis. LT biosynthesis is initiated by 5-lipoxygenase (5-LOX) with the assistance of the substrate-binding 5-LOX-activating protein at the nuclear membrane. Here, we contrast the structural and functional consequences of the binding of two natural product inhibitors of 5-LOX. The redox-type inhibitor nordihydroguaiaretic acid (NDGA) is lodged in the 5-LOX active site, now fully exposed by disordering of the helix that caps it in the apo-enzyme. In contrast, the allosteric inhibitor 3-acetyl-11-keto-beta-boswellic acid (AKBA) from frankincense wedges between the membrane-binding and catalytic domains of 5-LOX, some 30 Å from the catalytic iron. While enzyme inhibition by NDGA is robust, AKBA promotes a shift in the regiospecificity, evident in human embryonic kidney 293 cells and in primary immune cells expressing 5-LOX. Our results suggest a new approach to isoform-specific 5-LOX inhibitor development through exploitation of an allosteric site in 5-LOX.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Coordinates and structure factors (6N2W, 6NCF) are available at the Protein Bank, www.rcsb.org.
References
Haeggstrom, J. Z. & Funk, C. D. Lipoxygenase and leukotriene pathways: biochemistry, biology, and roles in disease. Chem. Rev. 111, 5866–5898 (2011).
Radmark, O., Werz, O., Steinhilber, D. & Samuelsson, B. 5-Lipoxygenase, a key enzyme for leukotriene biosynthesis in health and disease. Biochim Biophys. Acta 1851, 331–339 (2015).
Serhan, C. N. Pro-resolving lipid mediators are leads for resolution physiology. Nature 510, 92–101 (2014).
Shimizu, T. et al. Characterization of leukotriene A4 synthase from murine mast cells: evidence for its identity to arachidonate 5-lipoxygenase. Proc. Natl Acad. Sci. USA 83, 4175–4179 (1986).
Dixon, R. A. F. et al. Requirement of a 5-lipoxygenase-activating protein for leukotriene synthesis. Nature 343, 282–284 (1990).
Ferguson, A. D. et al. Crystal structure of inhibitor-bound human 5-lipoxygenase-activating protein. Science 317, 510–512 (2007).
Vickers, P. J., Deluca, C., Wong, E. & Abramovitz, M. The effect of 5-lipoxygenase-activating protein (FLAP) on substrate utilization by 5-lipoxygenase. Adv. Exp. Med Biol. 400A, 145–151 (1997).
Abramovitz, M. et al. 5-lipoxygenase-activating protein stimulates the utilization of arachidonic acid by 5-lipoxygenase. Eur. J. Biochem 215, 105–111 (1993).
Evans, J. F., Ferguson, A. D., Mosley, R. T. & Hutchinson, J. H. What’s all the FLAP about?: 5-lipoxygenase-activating protein inhibitors for inflammatory diseases. Trends Pharm. Sci. 29, 72–78 (2008).
Werz, O., Gerstmeier, J. & Garscha, U. Novel leukotriene biosynthesis inhibitors (2012-2016) as anti-inflammatory agents. Expert Opin. therapeutic Pat. 27, 607–620 (2017).
Pettersen, D., Davidsson, O. & Whatling, C. Recent advances for FLAP inhibitors. Bioorg. Med Chem. Lett. 25, 2607–2612 (2015).
Funk, C. D., Chen, X. S., Johnson, E. N. & Zhao, L. Lipoxygenase genes and their targeted disruption. Prostaglandins Other Lipid Mediat. 68-69, 303–312 (2002).
Schneider, C., Pratt, D. A., Porter, N. A. & Brash, A. R. Control of oxygenation in lipoxygenase and cyclooxygenase catalysis. Chem. Biol. 14, 473–488 (2007).
Brash, A. R. Lipoxygenases: occurrence, functions, catalysis, and acquisition of substrate. J. Biol. Chem. 274, 23679–23682 (1999).
Neau, D. B. et al. Crystal structure of a lipoxygenase in complex with substrate: the arachidonic acid-binding site of 8R-lipoxygenase. J. Biol. Chem. 289, 31905–31913 (2014).
Newcomer, M. E. & Brash, A. R. The structural basis for specificity in lipoxygenase catalysis. Protein Sci. 24, 298–309 (2015).
Gilbert, N. C. et al. The structure of human 5-lipoxygenase. Science 331, 217–219 (2011).
Bokoch, G. M. & Reed, P. W. Evidence for inhibition of leukotriene A4 synthesis by 5,8,11,14-eicosatetraynoic acid in guinea pig polymorphonuclear leukocytes. J. Biol. Chem. 256, 4156–4159 (1981).
Safayhi, H., Sailer, E. R. & Ammon, H. P. Mechanism of 5-lipoxygenase inhibition by acetyl-11-keto-beta-boswellic acid. Mol. Pharm. 47, 1212–1216 (1995).
Sailer, E. R., Schweizer, S., Boden, S. E., Ammon, H. P. & Safayhi, H. Characterization of an acetyl-11-keto-beta-boswellic acid and arachidonate-binding regulatory site of 5-lipoxygenase using photoaffinity labeling. Eur. J. Biochem 256, 364–368 (1998).
Poeckel, D. & Werz, O. Boswellic acids: biological actions and molecular targets. Curr. Med. Chem. 13, 3359–3369 (2006).
Abdel-Tawab, M., Werz, O. & Schubert-Zsilavecz, M. Boswellia serrata: an overall assessment of in vitro, preclinical, pharmacokinetic and clinical data. Clin. Pharmacokinet. 50, 349–369 (2011).
Sturner, K. H. et al. A standardised frankincense extract reduces disease activity in relapsing-remitting multiple sclerosis (the SABA phase IIa trial). J. Neurol. Neurosurg. Psychiatry 89, 330–338 (2017).
Werz, O. & Steinhilber, D. Development of 5-lipoxygenase inhibitors–lessons from cellular enzyme regulation. Biochem Pharm. 70, 327–333 (2005).
Kemal, C., Louis-Flamberg, P., Krupinski-Olsen, R. & Shorter, A. L. Reductive inactivation of soybean lipoxygenase 1 by catechols: a possible mechanism for regulation of lipoxygenase activity. Biochemistry 26, 7064–7072 (1987).
Mitra, S., Bartlett, S. G. & Newcomer, M. E. Identification of the substrate access portal of 5-lipoxygenase. Biochemistry 54, 6333–6342 (2015).
Schexnaydre, E. E. et al. A 5-lipoxygenase-specific sequence motif impedes enzyme activity and confers dependence on a partner protein. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1864, 543–551 (2018).
Ericsson, U. B., Hallberg, B. M., Detitta, G. T., Dekker, N. & Nordlund, P. Thermofluor-based high-throughput stability optimization of proteins for structural studies. Anal. Biochem 357, 289–298 (2006).
Eek, P. et al. Structure of a calcium-dependent 11R-lipoxygenase suggests a mechanism for Ca2+ regulation. J. Biol. Chem. 287, 22377–22386 (2012).
Rakonjac Ryge, M. et al. A mutation interfering with 5-lipoxygenase domain interaction leads to increased enzyme activity. Arch. Biochem Biophys. 545, 179–185 (2014).
Werz, O. Inhibition of 5-lipoxygenase product synthesis by natural compounds of plant origin. Planta Med. 73, 1331–1357 (2007).
Gerstmeier, J., Weinigel, C., Barz, D., Werz, O. & Garscha, U. An experimental cell-based model for studying the cell biology and molecular pharmacology of 5-lipoxygenase-activating protein in leukotriene biosynthesis. Biochim Biophys. Acta 1840, 2961–2969 (2014).
Werner, M. et al. Targeting biosynthetic networks of the proinflammatory and proresolving lipid metabolome. FASEB J. 33, 6140–6153 (2019).
Siemoneit, U. et al. On the interference of boswellic acids with 5-lipoxygenase: mechanistic studies in vitro and pharmacological relevance. Eur. J. Pharm. 606, 246–254 (2009).
Surette, M. E., Palmantier, R., Gosselin, J. & Borgeat, P. Lipopolysaccharides prime whole human blood and isolated neutrophils for the increased synthesis of 5-lipoxygenase products by enhancing arachidonic acid availability: involvement of the CD14 antigen. J. Exp. Med 178, 1347–1355 (1993).
Werz, O. et al. Human macrophages differentially produce specific resolvin or leukotriene signals that depend on bacterial pathogenicity. Nat. Commun. 9, 59 (2018).
Deng, B. et al. Maresin biosynthesis and identification of maresin 2, a new anti-inflammatory and pro-resolving mediator from human macrophages. PLoS ONE 9, e102362 (2014).
Carion, T. W. et al. Immunoregulatory role of 15-lipoxygenase in the pathogenesis of bacterial keratitis. FASEB J. 32, 5026–5038 (2018).
Sailer, E. R. et al. Acetyl-11-keto-beta-boswellic acid (AKBA): structure requirements for binding and 5-lipoxygenase inhibitory activity. Br. J. Pharm. 117, 615–618 (1996).
Gillmor, S. A., Villasenor, A., Fletterick, R., Sigal, E. & Browner, M. F. The structure of mammalian 15-lipoxygenase reveals similarity to the lipases and the determinants of substrate specificity. Nat. Struct. Biol. 4, 1003–1009 (1997); erratum 5, 242 (1998).
Choi, J., Chon, J. K., Kim, S. & Shin, W. Conformational flexibility in mammalian 15S-lipoxygenase: Reinterpretation of the crystallographic data. Proteins 70, 1023–1032 (2008).
Kobe, M. J., Neau, D. B., Mitchell, C. E., Bartlett, S. G. & Newcomer, M. E. The structure of human 15-lipoxygenase-2 with a substrate mimic. J. Biol. Chem. 289, 8562–8569 (2014).
Mandal, A. K. et al. The membrane organization of leukotriene synthesis. Proc. Natl Acad. Sci. USA 101, 6587–6592 (2004).
Mandal, A. K. et al. The nuclear membrane organization of leukotriene synthesis. Proc. Natl Acad. Sci. USA 105, 20434–20439 (2008).
Gerstmeier, J. et al. 5-Lipoxygenase-activating protein rescues activity of 5-lipoxygenase mutations that delay nuclear membrane association and disrupt product formation. FASEB J. 30, 1892–1900 (2016).
Neau, D. B. et al. The 1.85 A structure of an 8R-lipoxygenase suggests a general model for lipoxygenase product specificity. Biochemistry 48, 7906–7915 (2009).
Murphy, R. C. & Gijon, M. A. Biosynthesis and metabolism of leukotrienes. Biochem J. 405, 379–395 (2007).
Flamand, N., Luo, M., Peters-Golden, M. & Brock, T. G. Phosphorylation of serine 271 on 5-lipoxygenase and its role in nuclear export. J. Biol. Chem. 284, 306–313 (2009).
Laskowski, R. A. & Swindells, M. B. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 51, 2778–2786 (2011).
Pettersen, E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
Evans, P. R. & Murshudov, G. N. How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 69, 1204–1214 (2013).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Schweizer, S., Eichele, K., Ammon, H. P. & Safayhi, H. 3-Acetoxy group of genuine AKBA (acetyl-11-keto-beta-boswellic acid) is alpha-configurated. Planta Med. 66, 781–782 (2000).
Zwart, P. H. et al. Automated structure solution with the PHENIX suite. Methods Mol. Biol. 426, 419–435 (2008).
Dauter, Z., Li, M. & Wlodawer, A. Practical experience with the use of halides for phasing macromolecular structures: a powerful tool for structural genomics. Acta Crystallogr. D Biol. Crystallogr. 57, 239–249 (2001).
Parsons, S. Introduction to twinning. Acta Crystallogr. D Biol. Crystallogr. 59, 1995–2003 (2003).
Wang, C. K., Weeratunga, S. K., Pacheco, C. M. & Hofmann, A. DMAN: a Java tool for analysis of multi-well differential scanning fluorimetry experiments. Bioinformatics 28, 439–440 (2012).
Fischer, L., Szellas, D., Radmark, O., Steinhilber, D. & Werz, O. Phosphorylation- and stimulus-dependent inhibition of cellular 5-lipoxygenase activity by nonredox-type inhibitors. FASEB J. 17, 949–951 (2003).
Acknowledgements
This work was funded in part by grants to M.E.N. (nos. NIH HL107887 and AHA 16GRNT31000010, and the NIH P50AT002776 seed grant, and the Louisiana Governor’s Biotechnology Initiative) and O.W. (Deutsche Forschungsgemeinschaft (DFG, the German Research Foundation), project no. 316213987, SFB 1278 PolyTarget (project nos. A04 and C02), CRC 1127 ChemBioSys (project no. A04) and Free State of Thuringia and the European Social Fund (2016 FGR 0045)). J.G. received a Carl Zeiss postdoctoral stipend. Preliminary X-ray data were collected at the Center for Advanced Microstructures and Devices (Baton Rouge). We thank the staff at the Center for Advanced Microstructures and Devices for screening and data collection of macromolecular crystals at the Protein Crystallography beamline. The work is based on research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the National Institute of General Medical Sciences from the National Institutes of Health (grant no. P30 GM124165). The Eiger 16M detector on 24-ID-E beam line is funded by a NIH-ORIP HEI grant (no. S10OD021527). This research used resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under contract no. DE-AC02-06CH11357.
Author information
Authors and Affiliations
Contributions
M.E.N. and O.W. designed the study. N.C.G. designed and executed the protein expression, purification, biochemical evaluation of enzyme stability and crystallization, as well as crystal structure phasing, map interpretation and refinement. In addition, N.C.G. prepared the 5-LOX variants and executed the corresponding enzyme assays. D.B.N. collected and processed the diffraction data. E.E.S. contributed the initial LM production and immunofluorescence studies in HEK293 cells, and preliminary enzyme activity assays. J.G. performed the LM analysis in stably transfected HEK293 cells, neutrophils and M1-like MDM, provided the data evaluation and statistics and prepared the graphs. F.B. performed the immunofluorescence experiments and the analysis of SPM in HEK293 cells. U.G. analyzed the LM profile of isolated 5-LOX. M.E.N. and O.W. wrote the manuscript and all authors contributed to data interpretation and manuscript preparation.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Tables 1–4 and Figs. 1–10.
Rights and permissions
About this article
Cite this article
Gilbert, N.C., Gerstmeier, J., Schexnaydre, E.E. et al. Structural and mechanistic insights into 5-lipoxygenase inhibition by natural products. Nat Chem Biol 16, 783–790 (2020). https://doi.org/10.1038/s41589-020-0544-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-020-0544-7
This article is cited by
-
Lipoxygenase inhibitory synthetic derivatives of methyl gallate regulate gene expressions of COX-2 and cytokines to reduce animal model arthritis
Scientific Reports (2023)
-
Design, synthesis, and pharmacological evaluation of indazole carboxamides of N-substituted pyrrole derivatives as soybean lipoxygenase inhibitors
Molecular Diversity (2023)
-
Classic Phytochemical Antioxidant and Lipoxygenase Inhibitor, Nordihydroguaiaretic Acid, Activates Phospholipase D through Oxidant Signaling and Tyrosine Phosphorylation Leading to Cytotoxicity in Lung Vascular Endothelial Cells
Cell Biochemistry and Biophysics (2023)
-
Anti-inflammatory and Antinociceptive Potential of Verbascum latisepalum
Revista Brasileira de Farmacognosia (2022)
-
The Biological Activity of 3-O-Acetyl-11-keto-β-Boswellic Acid in Nervous System Diseases
NeuroMolecular Medicine (2022)