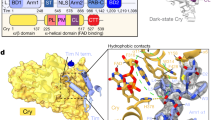
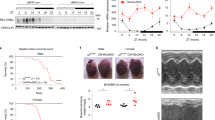
Abstract
CRY1 and CRY2 are essential components of the circadian clock controlling daily physiological rhythms. Accumulating evidences indicate distinct roles of these highly homologous proteins, in addition to redundant functions. Therefore, the development of isoform-selective compounds represents an effective approach towards understanding the similarities and differences of CRY1 and CRY2 by controlling each isoform individually. We conducted phenotypic screenings of circadian clock modulators, and identified KL101 and TH301 that selectively stabilize CRY1 and CRY2, respectively. Crystal structures of CRY–compound complexes revealed conservation of compound-binding sites between CRY1 and CRY2. We further discovered a unique mechanism underlying compound selectivity in which the disordered C-terminal region outside the pocket was required for the differential effects of KL101 and TH301 against CRY isoforms. By using these compounds, we found a new role of CRY1 and CRY2 as enhancers of brown adipocyte differentiation, providing the basis of CRY-mediated regulation of energy expenditure.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Bass, J. & Lazar, M. A. Circadian time signatures of fitness and disease. Science 354, 994–999 (2016).
Panda, S. Circadian physiology of metabolism. Science 354, 1008–1015 (2016).
Takahashi, J. S. Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 18, 164–179 (2017).
Chen, Z., Yoo, S. H. & Takahashi, J. S. Development and therapeutic potential of small-molecule modulators of circadian systems. Annu. Rev. Pharmacol. Toxicol. 58, 231–252 (2018).
Hirota, T. & Kay, S. A. Identification of small-molecule modulators of the circadian clock. Methods Enzymol. 551, 267–282 (2015).
Hirota, T. et al. Identification of small molecule activators of cryptochrome. Science 337, 1094–1097 (2012).
Xing, W. et al. SCF(FBXL3) ubiquitin ligase targets cryptochromes at their cofactor pocket. Nature 496, 64–68 (2013).
Nangle, S., Xing, W. & Zheng, N. Crystal structure of mammalian cryptochrome in complex with a small molecule competitor of its ubiquitin ligase. Cell Res. 23, 1417–1419 (2013).
van der Horst, G. T. et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature 398, 627–630 (1999).
Koike, N. et al. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 338, 349–354 (2012).
Huber, A. L. et al. CRY2 and FBXL3 cooperatively degrade c-MYC. Mol. Cell 64, 774–789 (2016).
Zhang, E. E. et al. Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nat. Med. 16, 1152–1156 (2010).
Lamia, K. A. et al. Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. Nature 480, 552–556 (2011).
Hirota, T. et al. High-throughput chemical screen identifies a novel potent modulator of cellular circadian rhythms and reveals CKIÉ‘ as a clock regulatory kinase. PLoS Biol. 8, e1000559 (2010).
Lee, J. W. et al. A small molecule modulates circadian rhythms through phosphorylation of the period protein. Angew. Chem. Int. Ed. Engl. 50, 10608–10611 (2011).
Oshima, T. et al. Cell-based screen identifies a new potent and highly selective CK2 inhibitor for modulation of circadian rhythms and cancer cell growth. Sci. Adv. 5, eaau9060 (2019).
Baggs, J. E. et al. Network features of the mammalian circadian clock. PLoS Biol. 7, e52 (2009).
Oshima, T. et al. C-H activation generates period-shortening molecules that target cryptochrome in the mammalian circadian clock. Angew. Chem. Int. Ed. Engl. 54, 7193–7197 (2015).
Lee, J. W. et al. Development of small-molecule cryptochrome stabilizer derivatives as modulators of the circadian clock. ChemMedChem 10, 1489–1497 (2015).
Czarna, A. et al. Structures of Drosophila cryptochrome and mouse cryptochrome1 provide insight into circadian function. Cell 153, 1394–1405 (2013).
Levy, C. et al. Updated structure of Drosophila cryptochrome. Nature 495, E3–E4 (2013).
Maul, M. J. et al. Crystal structure and mechanism of a DNA (6-4) photolyase. Angew. Chem. Int. Ed. Engl. 47, 10076–10080 (2008).
Chaves, I. et al. Functional evolution of the photolyase/cryptochrome protein family: importance of the C terminus of mammalian CRY1 for circadian core oscillator performance. Mol. Cell. Biol. 26, 1743–1753 (2006).
Gao, P. et al. Phosphorylation of the cryptochrome 1 C-terminal tail regulates circadian period length. J. Biol. Chem. 288, 35277–35286 (2013).
Sakakida, Y. et al. Importin É‘/β mediates nuclear transport of a mammalian circadian clock component, mCRY2, together with mPER2, through a bipartite nuclear localization signal. J. Biol. Chem. 280, 13272–13278 (2005).
Kurabayashi, N., Hirota, T., Sakai, M., Sanada, K. & Fukada, Y. DYRK1A and glycogen synthase kinase 3β, a dual-kinase mechanism directing proteasomal degradation of CRY2 for circadian timekeeping. Mol. Cell. Biol. 30, 1757–1768 (2010).
Patke, A. et al. Mutation of the human circadian clock gene CRY1 in familial delayed sleep phase disorder. Cell 169, 203–215.e13 (2017).
Rosensweig, C. et al. An evolutionary hotspot defines functional differences between CRYPTOCHROMES. Nat. Commun. 9, 1138 (2018).
Ohno, H., Shinoda, K., Spiegelman, B. M. & Kajimura, S. PPARγ agonists induce a white-to-brown fat conversion through stabilization of PRDM16 protein. Cell Metab. 15, 395–404 (2012).
Shinoda, K. et al. Genetic and functional characterization of clonally derived adult human brown adipocytes. Nat. Med. 21, 389–394 (2015).
Schmalen, I. et al. Interaction of circadian clock proteins CRY1 and PER2 is modulated by zinc binding and disulfide bond formation. Cell 157, 1203–1215 (2014).
Nangle, S. N. et al. Molecular assembly of the period-cryptochrome circadian transcriptional repressor complex. eLife 3, e03674 (2014).
Fribourgh, J. L. et al. Dynamics at the serine loop underlie differential affinity of cryptochromes for CLOCK:BMAL1 to control circadian timing. eLife 9, e55275 (2020).
Gerhart-Hines, Z. et al. The nuclear receptor Rev-erbÉ‘ controls circadian thermogenic plasticity. Nature 503, 410–413 (2013).
Nam, D. et al. The adipocyte clock controls brown adipogenesis through the TGF-β and BMP signaling pathways. J. Cell Sci. 128, 1835–1847 (2015).
Jordan, S. D. et al. CRY1/2 selectively repress pparδ and limit exercise capacity. Cell Metab. 26, 243–255 e6 (2017).
Yoo, S. H. et al. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc. Natl Acad. Sci. USA 101, 5339–5346 (2004).
Vitaterna, M. H. et al. Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc. Natl Acad. Sci. USA 96, 12114–12119 (1999).
Zhang, E. E. et al. A genome-wide RNAi screen for modifiers of the circadian clock in human cells. Cell 139, 199–210 (2009).
Liu, A. C. et al. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell 129, 605–616 (2007).
Ode, K. L. et al. Knockout-rescue embryonic stem cell-derived mouse reveals circadian-period control by quality and quantity of CRY1. Mol. Cell 65, 176–190 (2017).
Michael, A. K. et al. Formation of a repressive complex in the mammalian circadian clock is mediated by the secondary pocket of CRY1. Proc. Natl Acad. Sci. USA 114, 1560–1565 (2017).
Battye, T. G., Kontogiannis, L., Johnson, O., Powell, H. R. & Leslie, A. G. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 67, 271–281 (2011).
Winter, G. xia2: an expert system for macromolecular crystallography data reduction. J. Appl. Cryst. 43, 186–190 (2010).
Evans, P. Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 (2006).
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Murshudov, G. N. et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 (2011).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Acknowledgements
We thank N. Ono, Y. Niwa, A. Shiba, N. Kadofusa and Y. Aoki for technical assistance; T. Senda for technical advice; J.S. Takahashi (UT Southwestern) for Per2::Luc knock-in mice; T. Todo (Osaka University) for Cry1/Cry2 double knockout mice; and H.R. Ueda (University of Tokyo) for Cry1/Cry2 double knockout cells and pMU2-P(Cry1)-FLAG-I/RRE-Cry1 plasmid. This work was supported in part by JST PRESTO Grant No. JPMJPR14LA (T.H.); JSPS Grants No. 15H05590 and No. 18H02402 (T.H.); the Takeda Science Foundation (T.H.); the Suzuken Memorial Foundation (T.H.); AMED Grant No. JP19gm6110026 (M.H.); a JST PRESTO Grant (M.H.); JSPS Grants No. 16H06174, No. 19H03266 and No. 19K22693 (M.H.); the Uehara Memorial Foundation (M.H.); the Sumitomo Foundation (M.H.); the Astellas Foundation for Research on Metabolic Disorders (M.H.); the Cell Science Research Foundation (M.H.); JSPS Grant No. 18K06316 (Y.L.S.); and a MEXT PDIS Grant (S.O.). X-ray diffraction data collection and preliminary experiments were carried out at beamlines BL44XU of SPring-8 (proposals No. 2017A6743 and No. 2017B6743), BL41XU of SPring-8 (proposal No. 2018B1011) and BL-17A of the Photon Factory (proposals No. 2016R-63 and No. 2017G563). Recombinant CRY expression and beamline experiments were supported in part by BINDS from AMED (support No. JP19am0101074-0055 and No. JP19am0101071-0529).
Author information
Authors and Affiliations
Contributions
T.H. and M.H. conceptualized and administrated the project. S.M., Y.A., A. Srivastava and K.H. performed structural biology experiments. T.O. synthesized GO series compounds. Y.L.S. and M.H. conducted BAT experiments. E.M., Y.N., A. Sugiyama, A.H. and T.H. performed all other experiments. K.A., S.O. and A. Sato provided reagents and facilities. S.M., Y.L.S., Y.N., A. Srivastava, S.H., M.H. and T.H. validated data. T.H., M.H., S.M. and Y.L.S. visualized data and wrote the manuscript. T.H., M.H., S.M., Y.L.S., A. Srivastava, S.O. and S.H. edited the manuscript. All authors read and approved the manuscript. T.H., M.H., S.A.K., K.I. and F.T. supervised the project. T.H., M.H., Y.L.S. and S.O. acquired funding.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Tables 1–3, Figs. 1–10 and Note.
Rights and permissions
About this article
Cite this article
Miller, S., Son, Y.L., Aikawa, Y. et al. Isoform-selective regulation of mammalian cryptochromes. Nat Chem Biol 16, 676–685 (2020). https://doi.org/10.1038/s41589-020-0505-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-020-0505-1
This article is cited by
-
Targeted screening and identification of chlorhexidine as a pro-myogenic circadian clock activator
Stem Cell Research & Therapy (2023)
-
Molecular regulations of circadian rhythm and implications for physiology and diseases
Signal Transduction and Targeted Therapy (2022)
-
Discovery of a small molecule that selectively destabilizes Cryptochrome 1 and enhances life span in p53 knockout mice
Nature Communications (2022)
-
Live-cell imaging of circadian clock protein dynamics in CRISPR-generated knock-in cells
Nature Communications (2021)
-
Reversible modulation of circadian time with chronophotopharmacology
Nature Communications (2021)