Abstract
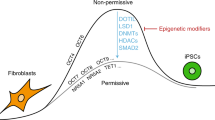
Silencing of the somatic cell type-specific genes is a critical yet poorly understood step in reprogramming. To uncover pathways that maintain cell identity, we performed a reprogramming screen using inhibitors of chromatin factors. Here, we identify acetyl-lysine competitive inhibitors targeting the bromodomains of coactivators CREB (cyclic-AMP response element binding protein) binding protein (CBP) and E1A binding protein of 300 kDa (EP300) as potent enhancers of reprogramming. These inhibitors accelerate reprogramming, are critical during its early stages and, when combined with DOT1L inhibition, enable efficient derivation of human induced pluripotent stem cells (iPSCs) with OCT4 and SOX2. In contrast, catalytic inhibition of CBP/EP300 prevents iPSC formation, suggesting distinct functions for different coactivator domains in reprogramming. CBP/EP300 bromodomain inhibition decreases somatic-specific gene expression, histone H3 lysine 27 acetylation (H3K27Ac) and chromatin accessibility at target promoters and enhancers. The master mesenchymal transcription factor PRRX1 is one such functionally important target of CBP/EP300 bromodomain inhibition. Collectively, these results show that CBP/EP300 bromodomains sustain cell-type-specific gene expression and maintain cell identity.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
RNA-sequencing, ChIP-sequencing and ATAC-sequencing data are deposited to the NCBI GEO database with the accession number GSE118220.
Code availability
The custom pipeline for ChIP-seq analysis can be found at: https://github.com/Acribbs/cribbslab/blob/master/Pipelines/pipeline_quantchip.py.
References
Takahashi, K. & Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006).
Xu, Y. et al. Transcriptional control of somatic cell reprogramming. Trends Cell Biol. 26, 272–288 (2016).
Papp, B. & Plath, K. Epigenetics of reprogramming to induced pluripotency. Cell 152, 1324–1343 (2013).
Mikkelsen, T. S. et al. Dissecting direct reprogramming through integrative genomic analysis. Nature 454, 49–55 (2008).
Huangfu, D. et al. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat. Biotechnol. 26, 795–797 (2008).
Onder, T. T. et al. Chromatin-modifying enzymes as modulators of reprogramming. Nature 483, 598–602 (2012).
Soufi, A., Donahue, G. & Zaret, K. S. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell 151, 994–1004 (2012).
Cheloufi, S. et al. The histone chaperone CAF-1 safeguards somatic cell identity. Nature 528, 218–224 (2015).
Zhuang, Q. et al. NCoR/SMRT co-repressors cooperate with c-MYC to create an epigenetic barrier to somatic cell reprogramming. Nat. Cell Biol. 20, 400–412 (2018).
Chronis, C. et al. Cooperative binding of transcription factors orchestrates reprogramming. Cell 168, 442–459.e20 (2017).
Ichida, J. K. et al. Notch inhibition allows oncogene-independent generation of iPS cells. Nat. Chem. Biol. 10, 632–639 (2014).
Jackson, S. A., Olufs, Z. P. G., Tran, K. A., Zaidan, N. Z. & Sridharan, R. Alternative routes to induced pluripotent stem cells revealed by reprogramming of the neural lineage. Stem Cell Rep. 6, 302–311 (2016).
Zhao, Y. et al. A XEN-like state bridges somatic cells to pluripotency during chemical reprogramming. Cell 163, 1678–1691 (2015).
Ogryzko, V. V., Schiltz, R. L., Russanova, V., Howard, B. H. & Nakatani, Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87, 953–959 (1996).
Delvecchio, M., Gaucher, J., Aguilar-Gurrieri, C., Ortega, E. & Panne, D. Structure of the p300 catalytic core and implications for chromatin targeting and HAT regulation. Nat. Struct. Mol. Biol. 20, 1040–1046 (2013).
Zeng, L., Zhang, Q., Gerona-Navarro, G., Moshkina, N. & Zhou, M.-M. Structural basis of site-specific histone recognition by the bromodomains of human coactivators PCAF and CBP/p300. Structure 16, 643–652 (2008).
Manning, E. T. T., Ikehara, T., Ito, T., Kadonaga, J. T. T. & Kraus, W. L. L. P300 forms a stable, template-committed complex with chromatin: role for the bromodomain. Mol. Cell. Biol. 21, 3876–3887 (2001).
Heintzman, N. D. et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39, 311–318 (2007).
Visel, A. et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457, 854–858 (2009).
Hnisz, D. et al. Super-enhancers in the control of cell identity and disease. Cell 155, 934–947 (2013).
Witte, S., Bradley, A., Enright, A. J. & Muljo, S. A. High-density P300 enhancers control cell state transitions. BMC Genomics 16, 903 (2015).
Iyer, N. G., Özdag, H. & Caldas, C. p300/CBP and cancer. Oncogene 23, 4225–4231 (2004).
Cribbs, A. et al. Inhibition of histone H3K27 demethylases selectively modulates inflammatory phenotypes of natural killer cells. J. Biol. Chem. 293, 2422–2437 (2018).
Arrowsmith, C. H. et al. The promise and peril of chemical probes. Nat. Chem. Biol. 11, 536–541 (2015).
Hay, D. A. et al. Discovery and optimization of small-molecule ligands for the CBP/p300 bromodomains. J. Am. Chem. Soc. 136, 9308–9319 (2014).
Picaud, S. et al. Generation of a selective small molecule inhibitor of the CBP/p300 bromodomain for leukemia therapy. Cancer Res. 75, 5106–5119 (2015).
Lasko, L. M. et al. Discovery of a selective catalytic p300/CBP inhibitor that targets lineage-specific tumours. Nature 550, 128–132 (2017).
Park, S. et al. Role of the CBP catalytic core in intramolecular SUMOylation and control of histone H3 acetylation. Proc. Natl Acad. Sci. USA 114, E5335–E5342 (2017).
Raisner, R. et al. Enhancer activity requires CBP/P300 bromodomain-dependent histone H3K27 acetylation. Cell Rep. 24, 1722–1729 (2018).
Weinert, B. T. et al. Time-resolved analysis reveals rapid dynamics and broad scope of the CBP/p300 acetylome. Cell 174, 231–244.e12 (2018).
Li, R. et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell 7, 51–63 (2010).
Nohno, T. et al. A chicken homeobox gene related to drosophila paired is predominantly expressed in the developing limb. Dev. Biol. 158, 254–264 (1993).
Lu, M. F. et al. prx-1 functions cooperatively with another paired-related homeobox gene, prx-2, to maintain cell fates within the craniofacial mesenchyme. Development 126, 495–504 (1999).
Shao, Z. et al. Reprogramming by de-bookmarking the somatic transcriptional program through targeting of BET bromodomains. Cell Rep. 16, 3138–3145 (2016).
Yao, T. P. et al. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell 93, 361–372 (1998).
Zhong, X. & Jin, Y. Critical roles of coactivator p300 in mouse embryonic stem cell differentiation and Nanog expression. J. Biol. Chem. 284, 9168–9175 (2009).
Fang, F. et al. Coactivators p300 and CBP maintain the identity of mouse embryonic stem cells by mediating long-range chromatin structure. Stem Cells 32, 1805–1816 (2014).
Cahan, P. et al. CellNet: network biology applied to stem cell engineering. Cell 158, 903–915 (2014).
Creyghton, M. P. et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl Acad. Sci. USA 107, 21931–21936 (2010).
Hammitzsch, A. et al. CBP30, a selective CBP/p300 bromodomain inhibitor, suppresses human Th17 responses. Proc. Natl Acad. Sci. USA 112, 10768–10773 (2015).
Conery, A. R. et al. Bromodomain inhibition of the transcriptional coactivators CBP/EP300 as a therapeutic strategy to target the IRF4 network in multiple myeloma. eLife 5, e10483 (2016).
Lynch, C. J. et al. The RNA polymerase II factor RPAP1 is critical for mediator-driven transcription and cell identity. Cell Rep. 22, 396–410 (2018).
Ortega, E. et al. Transcription factor dimerization activates the p300 acetyltransferase. Nature 562, 538–544 (2018).
Wang, W.-P. et al. The EP300, KDM5A, KDM6A and KDM6B chromatin regulators cooperate with klf4 in the transcriptional activation of POU5F1. PLoS One 7, e52556 (2012).
Merika, M., Williams, A. J., Chen, G., Collins, T. & Thanos, D. Recruitment of CBP/p300 by the IFN beta enhanceosome is required for synergistic activation of transcription. Mol. Cell 1, 277–287 (1998).
Simandi, Z. et al. OCT4 acts as an integrator of pluripotency and signal-induced differentiation. Mol. Cell 63, 647–661 (2016).
Ocaña, O. H. et al. Metastatic colonization requires the repression of the epithelial-mesenchymal transition inducer Prrx1. Cancer Cell 22, 709–724 (2012).
Du, B. et al. The transcription factor paired-related homeobox 1 (Prrx1) inhibits adipogenesis by activating transforming growth factor-β (TGFβ) signaling. J. Biol. Chem. 288, 3036–3047 (2013).
Yang, C. S., Lopez, C. G. & Rana, T. M. Discovery of nonsteroidal anti-inflammatory drug and anticancer drug enhancing reprogramming and induced pluripotent stem cell generation. Stem Cells 29, 1528–1536 (2011).
Park, I.-H. et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature 451, 141–146 (2008).
Seiler, C. Y. et al. DNASU plasmid and PSI:Biology-Materials repositories: resources to accelerate biological research. Nucleic Acids Res. 42, D1253–D1260 (2014).
Fidan, K. et al. Generation of integration-free induced pluripotent stem cells from a patient with familial Mediterranean fever (FMF). Stem Cell Res. 15, 694–696 (2015).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Buenrostro, J. D., Giresi, P. G., Zaba, L. C., Chang, H. Y. & Greenleaf, W. J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013).
Sims, D. et al. CGAT: computational genomics analysis toolkit. Bioinformatics 30, 1290–1291 (2014).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Acknowledgements
We would like to thank A. Kocabay and A.C. Taşkın for help with mouse experiments, A. Ruacan (Koç University, School of Medicine, Department of Pathology) for examination of histological sections and E. Hookway as well as T. Seker, E. Kavak (Genomize Inc.) for help with the initial generation and analyses of the RNA-seq data. The authors gratefully acknowledge use of the services and facilities of the Koç University Research Center for Translational Medicine (KUTTAM), funded by the Republic of Turkey Ministry of Development. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Ministry of Development. K.S. was supported by a TUBITAK BIDEB Scholarship. Work in the U.O. laboratory was supported by Arthritis Research UK (program grant no. 20522), Cancer Research UK and the Rosetrees Foundation. The Structural Genomics Consortium is a registered charity (no. 1097737) that receives funds from AbbVie, Bayer Pharma AG, Boehringer Ingelheim, Canada Foundation for Innovation, Eshelman Institute for Innovation, Genome Canada, Innovative Medicines Initiative (EU/EFPIA) (ULTRA-DD grant no. 115766), Janssen, Merck KGaA Darmstadt Germany, MSD, Novartis Pharma AG, Ontario Ministry of Economic Development and Innovation, Pfizer, São Paulo Research Foundation-FAPESP, Takeda, and Wellcome (106169/ZZ14/Z). The research has received funding from the People Programme (Marie Curie Actions) of the European Union’s Seventh Framework Programme (FP7/2007-2013) under REA grant agreement no. 609305. This work was supported by an EMBO Installation Grant (T.T.O.), a Newton Advanced Fellowship (U.O. and T.T.O.) and TUBITAK Project 213S182 (T.T.O.).
Author information
Authors and Affiliations
Contributions
A.E., K.S. and G.G.S. performed experiments and analyzed data. A.P.C. performed bioinformatics analyses. M.P. performed sequencing experiments. F.U. and T.M. analyzed RNA-seq data. J.E.D. provided materials. S.G. performed experiments. Ş.A. supervised research. U.O. designed the project, interpreted results and supervised the project. T.T.O. designed the project, interpreted results and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Tables 1 and 2, Supplementary Figures 1–7
Rights and permissions
About this article
Cite this article
Ebrahimi, A., Sevinç, K., Gürhan Sevinç, G. et al. Bromodomain inhibition of the coactivators CBP/EP300 facilitate cellular reprogramming. Nat Chem Biol 15, 519–528 (2019). https://doi.org/10.1038/s41589-019-0264-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-019-0264-z
This article is cited by
-
Group 3 medulloblastoma transcriptional networks collapse under domain specific EP300/CBP inhibition
Nature Communications (2024)
-
Streamlined quantitative analysis of histone modification abundance at nucleosome-scale resolution with siQ-ChIP version 2.0
Scientific Reports (2023)
-
A fast chemical reprogramming system promotes cell identity transition through a diapause-like state
Nature Cell Biology (2023)
-
AF10 (MLLT10) prevents somatic cell reprogramming through regulation of DOT1L-mediated H3K79 methylation
Epigenetics & Chromatin (2021)
-
Biological importance of OCT transcription factors in reprogramming and development
Experimental & Molecular Medicine (2021)