Abstract
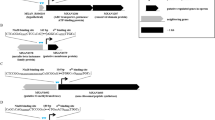
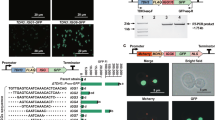
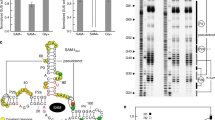
Here we report a transcription factor decoy strategy for targeted activation of eight large silent polyketide synthase and non-ribosomal peptide synthetase gene clusters, ranging from 50 to 134 kilobases (kb) in multiple streptomycetes, and characterization of a novel oxazole family compound produced by a 98-kb biosynthetic gene cluster. Owing to its simplicity and ease of use, this strategy can be scaled up readily for discovery of natural products in streptomycetes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
The DNA sequence encoding the oxazolepoxidomycin biosynthetic gene cluster from Streptomyces sp. NRRL F-4335 has been deposited to GenBank with accession code BK010686. All other data pertaining to this study are contained in the published article and its Supplementary Information files or are available from the corresponding author upon reasonable request.
References
Martens, E. & Demain, A. L. J. Antibiot. 70, 520–526 (2017).
Ikeda, H. et al. Nat. Biotechnol. 21, 526–531 (2003).
Bentley, S. D. et al. Nature 417, 141–147 (2002).
Ren, H., Wang, B. & Zhao, H. Curr. Opin. Biotechnol. 48, 21–27 (2017).
Nah, H.-J., Pyeon, H.-R., Kang, S.-H., Choi, S.-S. & Kim, E.-S. Front. Microbiol. 8, 394 (2017).
Rutledge, P. J. & Challis, G. L. Nat. Rev. Microbiol. 13, 509–523 (2015).
Hecker, M. & Wagner, A. H. Biochem. Pharmacol. 144, 29–34 (2017).
Gambari, R. Expert Opin. Ther. Pat. 21, 1755–1771 (2011).
Liu, G., Chater, K. F., Chandra, G., Niu, G. & Tan, H. Microbiol. Mol. Biol. Rev. 77, 112–143 (2013).
Kotake, C. et al. J. Antibiot. (Tokyo) 45, 1442–1450 (1992).
Chan, Y. A. et al. Proc. Natl. Acad. Sci. USA 103, 14349–14354 (2006).
Harunari, E., Komaki, H. & Igarashi, Y. Beilstein. J. Org. Chem. 13, 441–450 (2017).
Robertson, A. W. et al. J. Am. Chem. Soc. 138, 2200–2208 (2016).
Xu, G. et al. J. Biol. Chem. 285, 27440–27448 (2010).
Mori, T. et al. Tetrahedr. Lett. 26, 1073–1076 (1985).
Zhao, C. et al. J. Biol. Chem. 285, 20097–20108 (2010).
Helfrich, E. J. N. & Piel, J. Nat. Prod. Rep. 33, 231–316 (2016).
Wagner, D. T. et al. Structure 25, 1045–1055.e2 (2017).
Gay, D. C., Spear, P. J. & Keatinge-Clay, A. T. ACS Chem. Biol. 9, 2374–2381 (2014).
Shichijo, Y. et al. J. Am. Chem. Soc. 130, 12230–12231 (2008).
Gallimore, A. R. et al. Chem. Biol. 13, 453–460 (2006).
Zhang, M. M. et al. Nat. Chem. Biol. 13, 607–609 (2017).
Kieser, T., Bibb, M. J., Buttner, M. J., Chater, K. F. & Hopwood, D. A. Practical Streptomyces Genetics (John Innes Foundation, Norwich, 2000).
Gibson, D. G. et al. Nat. Methods 6, 343–345 (2009).
Acknowledgements
This work was supported by grant GM077596 (to H.Z.) from the US National Institutes of Health (NIH). Some of the data were collected in the Carl R. Woese Institute for Genomic Biology Core on a 600 MHz NMR funded by NIH grant number S10-RR028833, LC-MS at the MCB Metabolomics Center, and HRMS at the SCS Mass Spectrometry Laboratory. B.W. and F.G. dedicate this article to the memory of Keqian Yang, who made important contributions to their understanding of Streptomyces genetic regulation of secondary metabolism.
Author information
Authors and Affiliations
Contributions
B.W. and H.Z. conceived and designed the study, and wrote the manuscript. B.W. and F.G. performed the experiments and analysed the data, and purified compounds. B.W. and S.-H.D. analyzed the NMR data.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1–6, Supplementary Figures 1–10, Supplementary Notes 1 and 2
Rights and permissions
About this article
Cite this article
Wang, B., Guo, F., Dong, SH. et al. Activation of silent biosynthetic gene clusters using transcription factor decoys. Nat Chem Biol 15, 111–114 (2019). https://doi.org/10.1038/s41589-018-0187-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-018-0187-0
This article is cited by
-
Genetic toolbox for Photorhabdus and Xenorhabdus: pSEVA based heterologous expression systems and CRISPR/Cpf1 based genome editing for rapid natural product profiling
Microbial Cell Factories (2024)
-
Escherichia coli has an undiscovered ability to inhibit the growth of both Gram-negative and Gram-positive bacteria
Scientific Reports (2024)
-
Alternative therapeutic strategies to treat antibiotic-resistant pathogens
Nature Reviews Microbiology (2024)
-
Manipulation and epigenetic control of silent biosynthetic pathways in actinobacteria
World Journal of Microbiology and Biotechnology (2024)
-
A scalable platform to discover antimicrobials of ribosomal origin
Nature Communications (2022)