Abstract
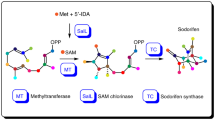
Terpene synthases typically form complex molecular scaffolds by concerted activation and cyclization of linear starting materials in a single enzyme active site. Here we show that iridoid synthase, an atypical reductive terpene synthase, catalyzes the activation of its substrate 8-oxogeranial into a reactive enol intermediate, but does not catalyze the subsequent cyclization into nepetalactol. This discovery led us to identify a class of nepetalactol-related short-chain dehydrogenase enzymes (NEPS) from catmint (Nepeta mussinii) that capture this reactive intermediate and catalyze the stereoselective cyclisation into distinct nepetalactol stereoisomers. Subsequent oxidation of nepetalactols by NEPS1 provides nepetalactones, metabolites that are well known for both insect-repellent activity and euphoric effects in cats. Structural characterization of the NEPS3 cyclase reveals that it binds to NAD+ yet does not utilize it chemically for a non-oxidoreductive formal [4 + 2] cyclization. These discoveries will complement metabolic reconstructions of iridoid and monoterpene indole alkaloid biosynthesis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The sequences of N. mussinii NEPS enzyme have been deposited in GenBank/EMBL/DDBJ with the accession codes MG677124 (NmNEPS1), MG677125 (NmNEPS2) and MG677126 (NmNEPS3). The NAD+ bound NmNEPS3 (7S-cis-cis-nepetalactol cyclase) X-ray structure has been deposited in the PDB with the accession code 6F9Q. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD008704. Detailed experimental procedures and can be found in the Supplementary Information. The authors declare that all other data supporting the findings of this study are available within this article and its Supplementary Information or from the authors upon reasonable request.
References
McElvain, S. M., Bright, R. D. & Johnson, P. R. The constituents of the volatile oil of catnip. I. Nepetalic acid, nepetalactone and related compounds. J. Am. Chem. Soc. 63, 1558–1563 (1941).
Formisano, C., Rigano, D. & Senatore, F. Chemical constituents and biological activities of Nepeta species. Chem. Biodivers. 8, 1783–1818 (2011).
Todd, N. B. Inheritance of the catnip response in domestic cats. J. Hered. 53, 54–56 (1962).
Bates, R. B. & Sigel, C. W. Terpenoids. cis-trans- and trans-cis- nepetalactones. Experientia 19, 564–565 (1963).
Waller, G. R., Price, G. H. & Mitchell, E. D. Feline attractant, cis,trans-nepetalactone: metabolism in the domestic cat. Science 164, 1281–1282 (1969).
Dawson, G. et al. Identification of an aphid sex pheromone. Nature 325, 614–616 (1987).
Clark, L. J., Hamilton, J. G. C., Chapman, J. V., Rhodes, M. J. C. & Hallahan, D. L. Analysis of monoterpenoids in glandular trichomes of the catmint Nepeta racemosa. Plant J. 11, 1387–1393 (1997).
Eisner, T. Catnip: its raison d’etre. Science 146, 1318–1320 (1964).
Birkett, M. A., Hassanali, A., Hoglund, S., Pettersson, J. & Pickett, J. A. Repellent activity of catmint, Nepeta cataria, and iridoid nepetalactone isomers against Afro-tropical mosquitoes, ixodid ticks and red poultry mites. Phytochemistry 72, 109–114 (2011).
Rajaonarivony, J. I. M., Gershenzon, J. & Croteau, R. Characterization and mechanism of (4 S)-limonene synthase, a monoterpene cyclase from the glandular trichomes of peppermint (Mentha x piperita). Arch. Biochem. Biophys. 296, 49–57 (1992).
Poulter, C. D., Argyle, J. C. & Mash, E. A. Letter: Prenyltransferase. New evidence for an ionization-condensation-elimination mechanism with 2-fluorogeranyl pyrophosphate. J. Am. Chem. Soc. 99, 957–959 (1977).
Baunach, M., Franke, J. & Hertweck, C. Terpenoid biosynthesis off the beaten track: unconventional cyclases and their impact on biomimetic synthesis. Angew. Chem. Int. Ed. Engl. 54, 2604–2626 (2015).
Tantillo, D. J. Importance of inherent substrate reactivity in enzyme-promoted carbocation cyclization/rearrangements. Angew. Chem. Int. Ed. Engl. 56, 10040–10045 (2017).
Miettinen, K. et al. The seco-iridoid pathway from Catharanthus roseus. Nat. Commun. 5, 3606 (2014).
Geu-Flores, F. et al. An alternative route to cyclic terpenes by reductive cyclization in iridoid biosynthesis. Nature 492, 138–142 (2012).
O’Connor, S. E. & Maresh, J. J. Chemistry and biology of monoterpene indole alkaloid biosynthesis. Nat. Prod. Rep. 23, 532–547 (2006).
Alagna, F. et al. Identification and characterization of the iridoid synthase involved in oleuropein biosynthesis in olive (Olea europaea) fruits. J. Biol. Chem. 291, 5542–5554 (2016).
Kries, H., Kellner, F., Kamileen, M. O. & O’Connor, S. E. Inverted stereocontrol of iridoid synthase in snapdragon. J. Biol. Chem. 292, 14659–14667 (2017).
Sherden, N. H. et al. Identification of iridoid synthases from Nepeta species: Iridoid cyclization does not determine nepetalactone stereochemistry. Phytochemistry 145, 48–56 (2018).
Kries, H. et al. Structural determinants of reductive terpene cyclization in iridoid biosynthesis. Nat. Chem. Biol. 12, 6–8 (2016).
Hu, Y. et al. Structures of iridoid synthase from Cantharanthus roseus with bound NAD+, NADPH, or NAD+/10-oxogeranial: reaction mechanisms. Angew. Chem. Int. Ed. Engl. 54, 15478–15482 (2015).
Qin, L. et al. Structure of iridoid synthase in complex with NADP+/8-oxogeranial reveals the structural basis of its substrate specificity. J. Struct. Biol. 194, 224–230 (2016).
Dawson, G. W., Pickett, J. A. & Smiley, D. W. M. The aphid sex pheromone cyclopentanoids: synthesis in the elucidation of structure and biosynthetic pathways. Bioorg. Med. Chem. 4, 351–361 (1996).
Liblikas, I. et al. Simplified isolation procedure and interconversion of the diastereomers of nepetalactone and nepetalactol.J. Nat. Prod. 68, 886–890 (2005).
Cucinotta, C. S., Ruini, A., Catellani, A. & Stirling, A. Ab initio molecular dynamics study of the keto-enol tautomerism of acetone in solution. Chemphyschem 7, 1229–1234 (2006).
Alagona, G., Ghio, C. & Nagy, P. I. The catalytic effect of water on the keto-enol tautomerism. Pyruvate and acetylacetone: a computational challenge. Phys. Chem. Chem. Phys. 12, 10173–10188 (2010).
Schreiber, S. L., Meyers, H. V. & Wiberg, K. B. Stereochemistry of the intramolecular enamine/enal (enone) cycloaddition reaction and subsequent Transformations. J. Am. Chem. Soc. 108, 8274–8277 (1986).
Hallahan, D. L., West, J. M., Smiley, D. W. M. & Pickett, J. A. Nepetalactol oxidoreductase in trichomes of the catmint Nepeta racemosa. Phytochemistry 48, 421–427 (1998).
Moummou, H., Kallberg, Y., Tonfack, L. B., Persson, B. & van der Rest, B. The plant short-chain dehydrogenase (SDR) superfamily: genome-wide inventory and diversification patterns. BMC Plant Biol. 12, 219 (2012).
Weng, J. K. & Noel, J. P. The remarkable pliability and promiscuity of specialized metabolism. Cold Spring Harb. Symp. Quant. Biol. 77, 309–320 (2012).
Tatsis, E. C. et al. A three enzyme system to generate the Strychnos alkaloid scaffold from a central biosynthetic intermediate. Nat. Commun. 8, 316 (2017).
Davin, L. B. et al. Stereoselective bimolecular phenoxy radical coupling by an auxiliary (dirigent) protein without an active center. Science 275, 362–366 (1997).
Pickel, B. et al. An enantiocomplementary dirigent protein for the enantioselective laccase-catalyzed oxidative coupling of phenols. Angew. Chem. Int. Ed. Engl. 49, 202–204 (2010).
Brown, S., Clastre, M., Courdavault, V. & O’Connor, S. E. De novo production of the plant-derived alkaloid strictosidine in yeast. Proc. Natl Acad. Sci. U.S.A. 112, 3205–3210 (2015).
Campbell, A. et al. Engineering of a nepetalactol-producing platform strain of Saccharomyces cerevisiae for the production of plant seco-iridoids. ACS Synth. Biol. 5, 405–414 (2016).
Billingsley, J. M. et al. Engineering the biocatalytic selectivity of iridoid production in Saccharomyces cerevisiae. Metab. Eng. 44, 117–125 (2017).
Tyanova, S., Temu, T. & Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 11, 2301–2319 (2016).
Kabsch, W. XDS Acta Crystallogr. D. Biol. Crystallogr. 66, 125–132 (2010).
Winter, G. Xia2: An expert system for macromolecular crystallography data reduction. J. Appl. Crystallogr. 43, 186–190 (2010).
Evans, P. R. & Murshudov, G. N. How good are my data and what is the resolution? Acta Crystallogr. D. Biol. Crystallogr. 69, 1204–1214 (2013).
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D. Biol. Crystallogr. 67, 235–242 (2011).
Youn, B., Moinuddin, S. G. A., Davin, L. B., Lewis, N. G. & Kang, C. Crystal structures of apo-form and binary/ternary complexes of Podophyllum secoisolariciresinol dehydrogenase, an enzyme involved in formation of health-protecting and plant defense lignans. J. Biol. Chem. 280, 12917–12926 (2005).
Bunkóczi, G. & Read, R. J. Improvement of molecular-replacement models with Sculptor. Acta Crystallogr. D. Biol. Crystallogr. 67, 303–312 (2011).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Cowtan, K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D. Biol. Crystallogr. 62, 1002–1011 (2006).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 66, 486–501 (2010).
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D. Biol. Crystallogr. 53, 240–255 (1997).
Davis, I. W. et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 (2007).
Zhang, Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9, 40 (2008).
Krieger, E. et al. Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: four approaches that performed well in CASP8. Proteins 77, 114–122 (2009).
Trott, O. & Olson, A. J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 (2010).
Larkin, M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007).
Robert, X. & Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 (2014).
Mint Evolutionary Genomics Consortium. Phylogenomic mining of the mints reveals multiple mechanisms contributing to the evolution of chemical diversity in Lamiaceae. Mol. Plant 11, 1084–1096 (2018).
Ringer, K. L., Davis, E. M. & Croteau, R. Monoterpene metabolism. Cloning, expression, and characterization of (-)-isopiperitenol/(-)-carveol dehydrogenase of peppermint and spearmint. Plant Physiol. 137, 863–872 (2005).
Edgar, R. C. Muscle: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5, 113 (2004).
Nguyen, L. T., Schmidt, H. A., von Haeseler, A. & Minh, B. Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015).
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A. & Jermiin, L. S. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589 (2017).
Minh, B. Q., Nguyen, M. A. T. & von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 30, 1188–1195 (2013).
Acknowledgements
We acknowledge funding from UK Biotechnological and Biological Sciences Research Council (BBSRC) and Engineering and Physical Sciences Research Council (EPSRC) joint-funded OpenPlant Synthetic Biology Research Centre (BB/L014130/1) and from the National Science Foundation Plant Genome Research Program (IOS- 1444499). For the X-ray data collection, we acknowledge Diamond Light Source for access to beamline I03 under proposal MX13467, with support from the European Community’s Seventh Framework Program (FP7/2007–2013) under grant agreement 283570 (BioStruct-X). We are grateful to: P. Brett (John Innes Centre) for assistance with GC–MS analysis and M. Vigoroux (John Innes Centre) for assistance with proteome annotations. We also thank K. Houk, J. Fell, H. Kries and D. Whitaker for discussions concerning the iridoid synthase and cyclization mechanisms.
Author information
Authors and Affiliations
Contributions
S.E.O.’C. designed and supervised the project; B.R.L. performed molecular cloning, protein purification, enzyme assays, trichome isolation, chemical synthesis, phylogenetic analysis, homology modeling and computational docking; G.S. performed proteome analysis; M.O.K. assisted with protein purification, compound isolation and chemical synthesis; B.R.L., G.R.T. and C.E.M.S. performed crystallization trials and obtained crystals; B.R.L., G.R.T. and D.M.L. refined structures; B.R.L. and S.E.O.’C. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
A UK patent application has been submitted based on the work reported here (GB1808663.7).
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Tables 1–4, Supplementary Figures 1–17
Supplementary Note 1
Synthetic Procedures
Supplementary Data Set 1
Proteomic Data
Rights and permissions
About this article
Cite this article
Lichman, B.R., Kamileen, M.O., Titchiner, G.R. et al. Uncoupled activation and cyclization in catmint reductive terpenoid biosynthesis. Nat Chem Biol 15, 71–79 (2019). https://doi.org/10.1038/s41589-018-0185-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-018-0185-2
This article is cited by
-
Characterization of the horse chestnut genome reveals the evolution of aescin and aesculin biosynthesis
Nature Communications (2023)
-
Biosynthesis, natural distribution, and biological activities of acyclic monoterpenes and their derivatives
Phytochemistry Reviews (2023)
-
Reconstitution of monoterpene indole alkaloid biosynthesis in genome engineered Nicotiana benthamiana
Communications Biology (2022)
-
Biocatalytic routes to stereo-divergent iridoids
Nature Communications (2022)
-
In vivo characterization of key iridoid biosynthesis pathway genes in catnip (Nepeta cataria)
Planta (2022)